3DYO
 
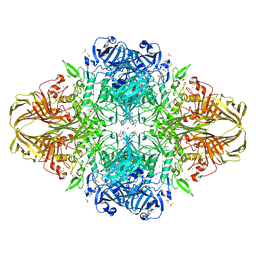 | | E. coli (lacZ) beta-galactosidase (H418N) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H, Huber, R.E, Matthews, B.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
3FBY
 
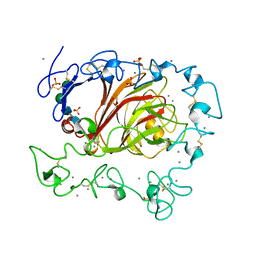 | |
5U4I
 
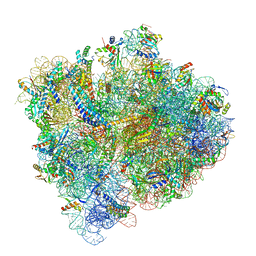 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
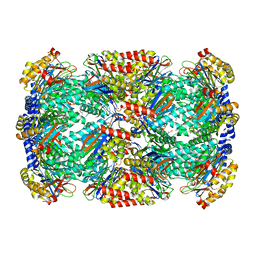
3H6F
 
 | |
5V8I
 
 | | Thermus thermophilus 70S ribosome lacking ribosomal protein uS17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Gregory, S.T, Jogl, G. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
To be published
|
|
5V2C
 
 | | RE-REFINEMENT OF CRYSTAL STRUCTURE OF PHOTOSYSTEM II COMPLEX | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, J, Wiwczar, J.M, Brudvig, G.W. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlorophyll a with a farnesyl tail in thermophilic cyanobacteria.
Photosyn. Res., 134, 2017
|
|
5VPP
 
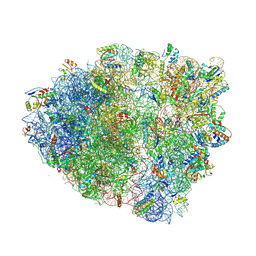 | | The 70S P-site tRNA SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VYC
 
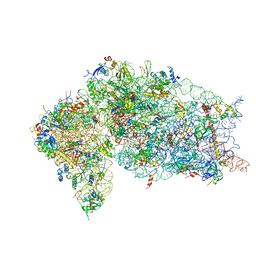 | | Crystal structure of the human 40S ribosomal subunit in complex with DENR-MCT-1. | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Stolboushkina, E.A, Vaidya, A.T, Garber, M.B, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structure of the Human Ribosome in Complex with DENR-MCT-1.
Cell Rep, 20, 2017
|
|
5VPO
 
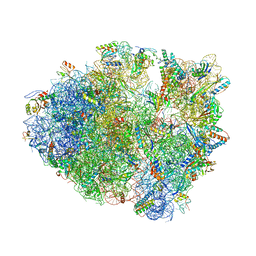 | | The 70S P-site ASL SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3H6I
 
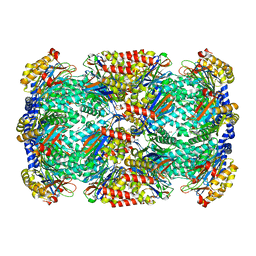 | |
3EU8
 
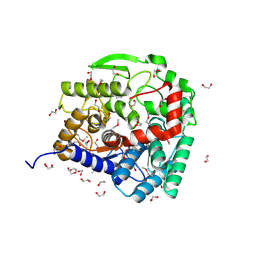 | |
3HHQ
 
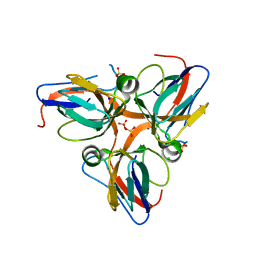 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3HFA
 
 | |
3HF9
 
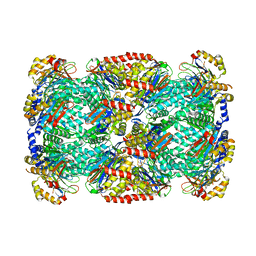 | |
3I3D
 
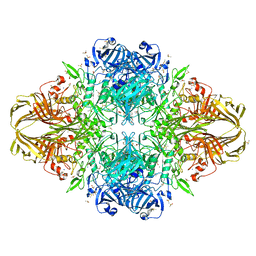 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
3I55
 
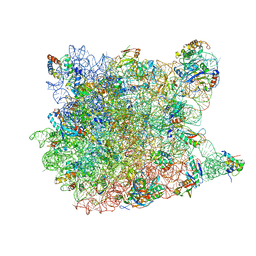 | | Co-crystal structure of Mycalamide A Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
1Z8G
 
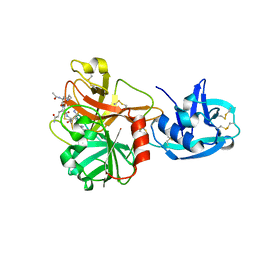 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
1YI2
 
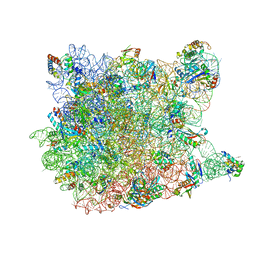 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-11 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YHQ
 
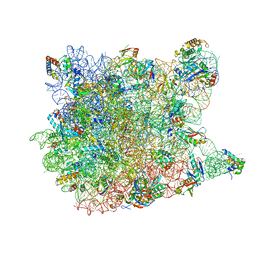 | | Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIT
 
 | | Crystal Structure Of Virginiamycin M and S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIJ
 
 | | Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-12 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJN
 
 | | Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJW
 
 | | Crystal Structure Of Quinupristin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-15 | | Release date: | 2005-04-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1XMO
 
 | | Crystal Structure of mnm5U34t6A37-tRNALysUUU Complexed with AAG-mRNA in the Decoding Center | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
