7OQ4
 
 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
4I43
 
 | | Crystal structure of Prp8:Aar2 complex | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Galej, W.P, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2012-11-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Prp8 reveals active site cavity of the spliceosome.
Nature, 493, 2013
|
|
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
2FJB
 
 | | Adenosine-5'-phosphosulfate reductase im complex with products | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, ADENOSINE MONOPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
7Q4O
 
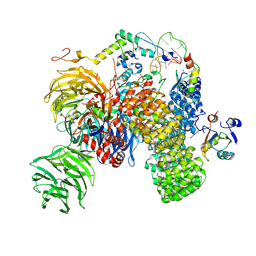 | | Substrate-bound A-like U2 snRNP | | Descriptor: | BPS oligo, PHD finger-like domain-containing protein 5A, Splicing factor 3A subunit 2, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-11-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
7Q4P
 
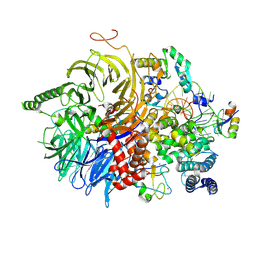 | | U2 snRNP after ATP-dependent remodelling | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3A subunit 2, Splicing factor 3A subunit 3, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-11-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
7Q3L
 
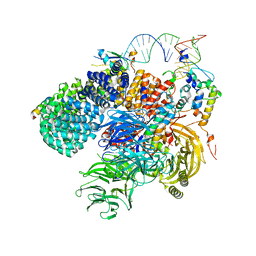 | | Human 17S U2 snRNP 5' domain | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
7PYT
 
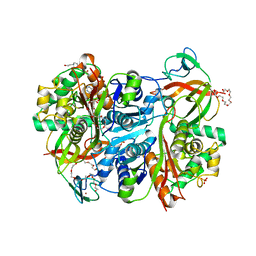 | | Benzoylsuccinyl-CoA thiolase with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Benzoylsuccinyl-CoA thiolase subunit, ... | | Authors: | Ermler, U, Heider, J, Weidenweber, S, Demmer, U. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
7PXP
 
 | | Benzoylsuccinyl-CoA thiolase | | Descriptor: | Benzoylsuccinyl-CoA thiolase subunit, ZINC ION | | Authors: | Ermler, U, Heider, H, Weidenweber, S, Demmer, U. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
2FHZ
 
 | |
2FJE
 
 | | adenosine-5-phosphosulfate reductase oxidized state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, adenylylsulfate reductase, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2FJD
 
 | | adenosine-5-phosphosulfate reductase in complex with sulfite (covalent adduct) | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, IRON/SULFUR CLUSTER, adenylylsulfate reductase, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
4ILG
 
 | | Crystal structure of Aar2p in complex with the Prp8p RNaseH and Jab1/MPN domains | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weber, G, Heroven, A.C, Santos, K.F, Wahl, M.C. | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for dual roles of Aar2p in U5 snRNP assembly.
Genes Dev., 27, 2013
|
|
4ILH
 
 | |
2FJA
 
 | | adenosine 5'-phosphosulfate reductase in complex with substrate | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2GOX
 
 | | Crystal structure of Efb-C / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-04-14 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural basis for complement inhibition by Staphylococcus aureus.
Nat.Immunol., 8, 2007
|
|
5YBB
 
 | | Structural basis underlying complex assembly andconformational transition of the type I R-M system | | Descriptor: | DNA, Restriction endonuclease S subunits, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, Y.P, Tang, Q, Zhang, J.Z, Tian, L.F, Gao, P, Yan, X.X. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis underlying complex assembly and conformational transition of the type I R-M system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YLZ
 
 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
5Y88
 
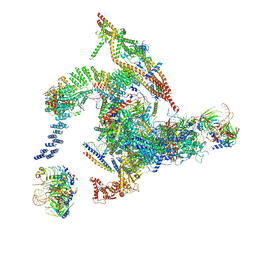 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5WSG
 
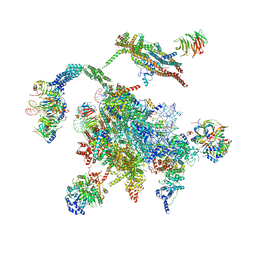 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | Descriptor: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
5XJC
 
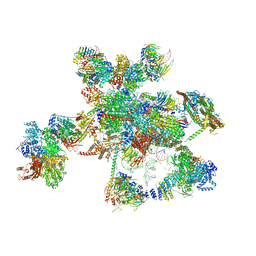 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
7BTO
 
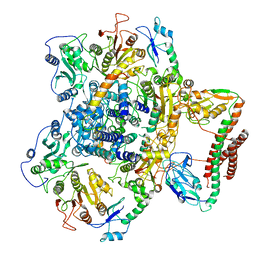 | | EcoR124I-ArdA in the Translocation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTP
 
 | | EcoR124I-Ocr in Restriction-Alleviation State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
