7D0T
 
 | |
4Q0B
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-04-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PWD
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with bulge-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*U)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4ALD
 
 | | HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-BISPHOSPHATE ALDOLASE | | Authors: | Dalby, A.R, Dauter, Z, Littlechild, J.A. | | Deposit date: | 1998-07-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications.
Protein Sci., 8, 1999
|
|
5UGC
 
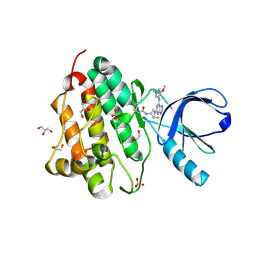 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
4DG1
 
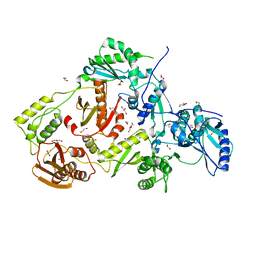 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
3PMO
 
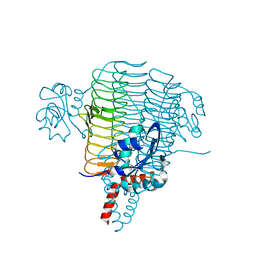 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3G2L
 
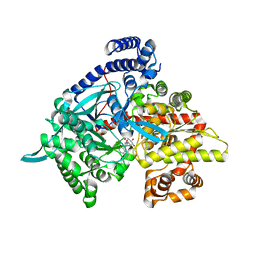 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | 1-beta-D-glucopyranosyl-4-naphthalen-1-yl-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
7CVR
 
 | | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine | | Descriptor: | (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2S)-3-phenyl-2-[[(2S)-1-phenylmethoxycarbonylpyrrolidin-2-yl]carbonylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine at 1.60 Angstrom Resolution
To Be Published
|
|
6AFI
 
 | | DJ-1 with compound 11 | | Descriptor: | 1-ethylindole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFG
 
 | | DJ-1 with compound 9 | | Descriptor: | 1-methylindole-2,3-dione, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
6AFH
 
 | | DJ-1 with compound 10 | | Descriptor: | 1-(2-phenylethyl)indole-2,3-dione, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
5CSU
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarviostatin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
4O0T
 
 | |
3MHG
 
 | | Dihydroxyacetone phosphate carbanion intermediate in tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, CALCIUM ION, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | LowKam, C, Liotard, B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity.
J.Biol.Chem., 285, 2010
|
|
3IS9
 
 | | Crystal structure of the HIV-1 reverse transcriptase (RT) in complex with the alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate). | | Descriptor: | Reverse transcriptase, Reverse transcriptase/ribonuclease H, dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate) | | Authors: | Ho, W.C, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2009-08-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic study of a novel subnanomolar inhibitor provides insight on the binding interactions of alkenyldiarylmethanes with human immunodeficiency virus-1 reverse transcriptase.
J.Med.Chem., 52, 2009
|
|
5UG9
 
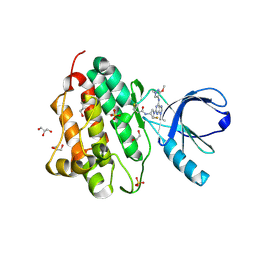 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
7WJE
 
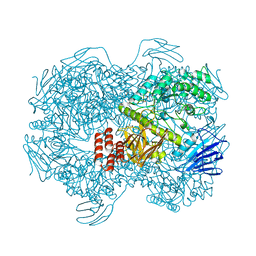 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotetraose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
4TPP
 
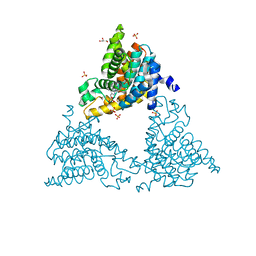 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
4LSU
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG20 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
3G2K
 
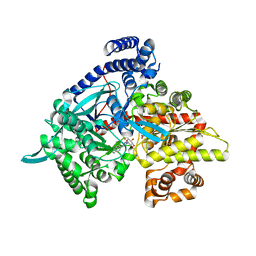 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazole | | Descriptor: | 1-beta-D-glucopyranosyl-4-naphthalen-2-yl-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
3GPB
 
 | |
1J9W
 
 | | Solution Structure of the CAI Michigan 1 Variant | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, ZINC ION | | Authors: | Briganti, F, Ferraroni, M, Chedwiggen, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination.
Biochemistry, 41, 2002
|
|
1JV0
 
 | | THE CRYSTAL STRUCTURE OF THE ZINC(II) ADDUCT OF THE CAI MICHIGAN 1 VARIANT | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, CHLORIDE ION, ... | | Authors: | Briganti, F, Ferraroni, M, Chegwidden, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination
Biochemistry, 41, 2002
|
|
3NBP
 
 | | HIV-1 reverse transcriptase with aminopyrimidine inhibitor 2 | | Descriptor: | 4-(4-{[4-(4-cyano-2,6-dimethylphenoxy)pyrimidin-2-yl]amino}piperidin-1-yl)benzenesulfonamide, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
