3D9K
 
 | |
3D9I
 
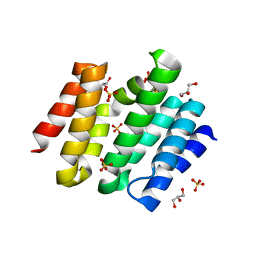 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, GLYCEROL, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9O
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9L
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | ACETATE ION, CTD-PEPTIDE, GLYCEROL, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9P
 
 | |
2F21
 
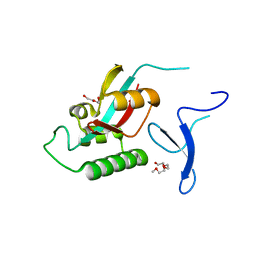 | | human Pin1 Fip mutant | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Bowman, M.E, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-11-15 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7CEI
 
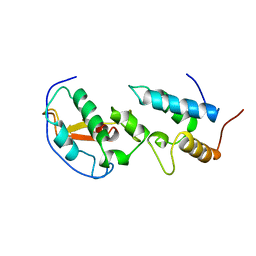 | | THE ENDONUCLEASE DOMAIN OF COLICIN E7 IN COMPLEX WITH ITS INHIBITOR IM7 PROTEIN | | Descriptor: | PROTEIN (COLICIN E7 IMMUNITY PROTEIN), ZINC ION | | Authors: | Ko, T.P, Liao, C.C, Ku, W.Y, Chak, K.F, Yuan, H.S. | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Structure Fold.Des., 7, 1999
|
|
4FXJ
 
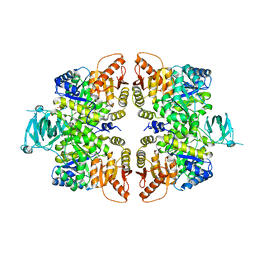 | |
2M8J
 
 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M9J
 
 | | NMR solution structure of Pin1 WW domain mutant 6-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
7MDC
 
 | | Full-length wildtype ClbP inhibited by hexanoyl-D-asparagine boronic acid | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Velilla, J.A, Volpe, M.R, Gaudet, R. | | Deposit date: | 2021-04-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A small molecule inhibitor prevents gut bacterial genotoxin production.
Nat.Chem.Biol., 19, 2023
|
|
7P71
 
 | | The PDZ domain of MAGI1_2 complexed with the PDZ-binding motif of HPV35-E6 | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
2M9F
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M16
 
 | | P75/LEDGF PWWP Domain | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2012-11-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes.
Nucleic Acids Res., 41, 2013
|
|
2M8I
 
 | | Structure of Pin1 WW domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9I
 
 | | NMR solution structure of Pin1 WW domain variant 6-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2MSR
 
 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
2MTN
 
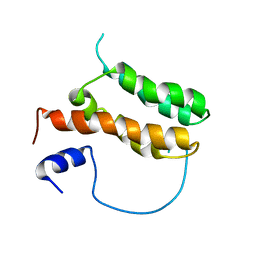 | | Solution structure of MLL-IBD complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein fusion | | Authors: | Cierpicki, T, Pollock, J, Murai, M. | | Deposit date: | 2014-08-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The same site on the integrase-binding domain of lens epithelium-derived growth factor is a therapeutic target for MLL leukemia and HIV.
Blood, 124, 2014
|
|
2N3A
 
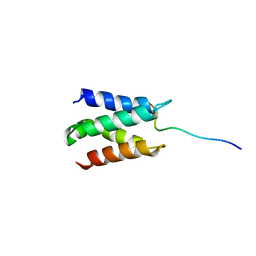 | | Solution structure of LEDGF/p75 IBD in complex with POGZ peptide (1389-1404) | | Descriptor: | PC4 and SFRS1-interacting protein, Pogo transposable element with ZNF domain | | Authors: | Tesina, P, Cermakova, K, Horejsi, M, Prochazkova, K, Fabry, M, Sharma, S, Christ, F, Demeulemeester, J, Debyser, Z, De Rijck, J, Veverka, V, Rezacova, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif.
Nat Commun, 6, 2015
|
|
7C36
 
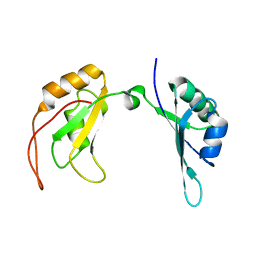 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
2N64
 
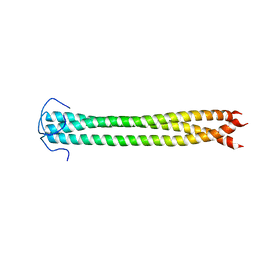 | |
8WWY
 
 | | Crystal structure of mouse TIFA/TIFAB heterodimer | | Descriptor: | (2R,5R)-hexane-2,5-diol, TRAF-interacting protein with FHA domain-containing protein A, TRAF-interacting protein with FHA domain-containing protein B | | Authors: | Nakamura, T. | | Deposit date: | 2023-10-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | TIFAB regulates the TIFA-TRAF6 signaling pathway involved in innate immunity by forming a heterodimer complex with TIFA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ETJ
 
 | | C5 portal vertex in the partially-enveloped virion capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
7ETO
 
 | | C1 CVSC-binding penton vertex in the virion capsid of Human Cytomegalovirus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Nat Commun, 12, 2021
|
|
