9IP2
 
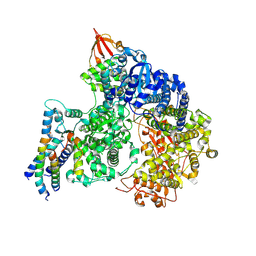 | | Cryo-EM structure of the RNA-dependent RNA polymerase complex from Marburg virus | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Polymerase cofactor VP35, RNA-directed RNA polymerase L,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Li, G, Du, T, Wang, J, Wu, S, Ru, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the RNA-dependent RNA polymerase complexes from highly pathogenic Marburg and Ebola viruses.
Nat Commun, 16, 2025
|
|
2WIJ
 
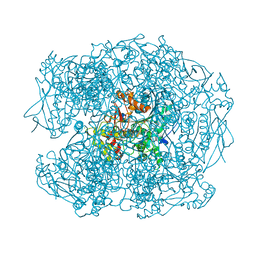 | | NONAGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
9I76
 
 | |
6TJT
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide | | Descriptor: | 1,2-ETHANEDIOL, Neuropilin-2, SULFATE ION, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFC peptide
To Be Published
|
|
8S0O
 
 | | A fragment-based inhibitor of SHP2 | | Descriptor: | 3-[4-chloranyl-2-(1H-pyrazol-4-ylmethyl)indazol-5-yl]-5-methyl-6-(piperazin-1-ylmethyl)-1H-pyrrolo[3,2-b]pyridine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
6ENS
 
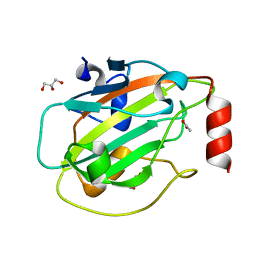 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9IP3
 
 | | Cryo-EM structure of the RNA-dependent RNA polymerase complex in a compact conformation from Ebola virus | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Polymerase cofactor VP35, Maltose/maltodextrin-binding periplasmic protein,RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, G, Du, T, Wang, J, Wu, S, Ru, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the RNA-dependent RNA polymerase complexes from highly pathogenic Marburg and Ebola viruses.
Nat Commun, 16, 2025
|
|
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
3IAI
 
 | | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 9, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4XX0
 
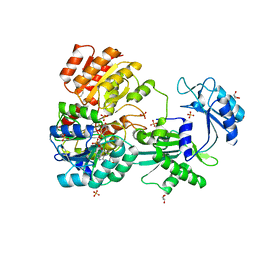 | | CoA bound to pig GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fraser, M.E, Huang, J, Malhi, M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of GTP-specific succinyl-CoA synthetase in complex with CoA.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8S0Q
 
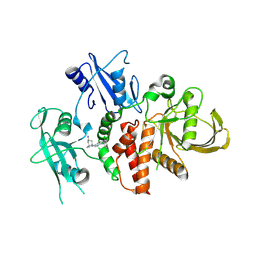 | | A fragment-based inhibitor of SHP2 | | Descriptor: | (1S,5R)-8-[3-[2,3-bis(chloranyl)phenyl]-1H-pyrrolo[3,2-b]pyridin-6-yl]-8-azabicyclo[3.2.1]octan-3-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Cleasby, A, Price, A. | | Deposit date: | 2024-02-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
5C0R
 
 | | Crystal Structure of a Generation 3 Influenza Hemagglutinin Stabilized Stem Complexed with the Broadly Neutralizing Antibody C179 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C179 Fab heavy chain, ... | | Authors: | Boyington, J.C, kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
2A74
 
 | | Human Complement Component C3c | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Complement Component C3c, GLYCEROL, ... | | Authors: | Janssen, B.J.C, Huizinga, E.G, Raaijmakers, H.C.A, Roos, A, Daha, M.R, Nilsson-Ekdahl, K, Nilsson, B, Gros, P. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of complement component C3 provide insights into the function and evolution of immunity.
Nature, 437, 2005
|
|
5C1M
 
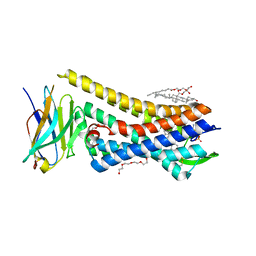 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
4AA1
 
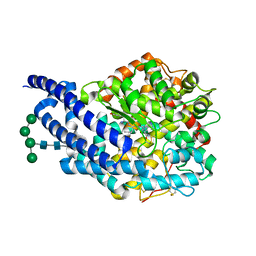 | | Crystal structure of ANCE in complex with Angiotensin-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-2, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Isaac, R.E, Akif, M, Schwager, S.L.U, Masuyer, G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-11-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
4YQ1
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | N-(1-{[(3-tert-butylbenzoyl)amino]methyl}cyclohexyl)-2,1-benzoxazole-4-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | To Be Published
To be published
|
|
7EVQ
 
 | | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Viswanathan, V, Singh, J, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution
To Be Published
|
|
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
4YQR
 
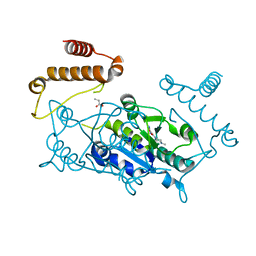 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)(4-methylpiperazin-1-yl)methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
5IDY
 
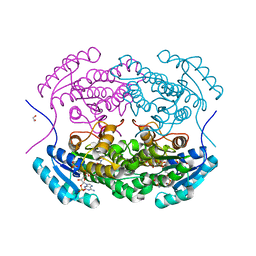 | |
6HOP
 
 | |
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
5FSM
 
 | | MTH1 substrate recognition: Complex with a methylbenzimidazolyl acetamide. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
6EAC
 
 | | Pseudomonas syringae SelO | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2018-08-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Protein AMPylation by an Evolutionarily Conserved Pseudokinase.
Cell, 175, 2018
|
|
