6SL8
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
1CAQ
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | Descriptor: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-23 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
5ZFH
 
 | | Mouse Kallikrein 7 | | Descriptor: | Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
7AK0
 
 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
4NZW
 
 | | Crystal Structure of STK25-MO25 Complex | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | Authors: | Feng, M, Hao, Q, Zhou, Z.C. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4Y8R
 
 | | Yeast 20S proteasome beta2-H116D mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4Y9V
 
 | | Gp54 tailspike of Acinetobacter baumannii bacteriophage AP22 in complex with A. baumannii capsular saccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2,4-dideoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-mannopyranuronic acid, CHLORIDE ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2015-02-17 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of Acinetobacter baumannii bacteriophage AP22 polysaccharide degrading lyase in complex with A. baumannii capsular saccharide at 0.9 A resolution
TO BE PUBLISHED
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
6EP9
 
 | |
8THJ
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
1BQ0
 
 | |
5K42
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with GDP-glucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
9J0F
 
 | | NADH bound purinergic receptor P2Y14 in complex with Gi | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Gu, Q.C, Tang, W.Q. | | Deposit date: | 2024-08-02 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural insights into ligand recognition and activation of human purinergic receptor P2Y14.
Cell Discov, 11, 2025
|
|
4OYA
 
 | |
8Z6X
 
 | |
6V01
 
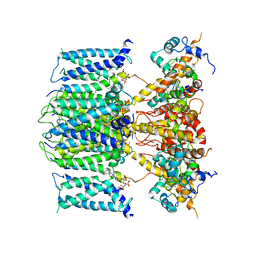 | | structure of human KCNQ1-KCNE3-CaM complex with PIP2 | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily E member 3, ... | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-18 | | Release date: | 2019-12-04 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
6V0P
 
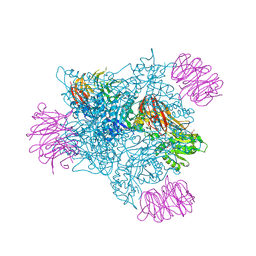 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
3MCQ
 
 | |
6C9F
 
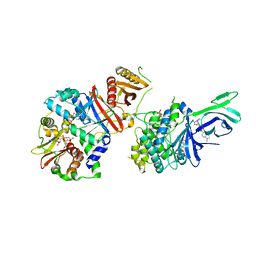 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
7G7F
 
 | | Crystal Structure of rat Autotaxin in complex with 1H-benzotriazol-5-yl-[rac-(3aR,8aS)-6-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-1,3,3a,4,5,7,8,8a-octahydropyrrolo[3,4-d]azepin-2-yl]methanone, i.e. SMILES C1C[C@@H]2[C@H](CCN1C(=O)c1cc(nc(c1)OCC1CCOCC1)C1CC1)CN(C2)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.00129469 microM | | Descriptor: | ACETATE ION, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G57
 
 | | Crystal Structure of rat Autotaxin in complex with (3,5-dichlorophenyl)methyl rac-(3aR,6aS)-5-(1H-triazol-5-ylmethylcarbamoyl)-3,3a,4,5,6,6a-hexahydro-1H-cyclopenta[c]pyrrole-2-carboxylate, i.e. SMILES [C@H]12CN(C[C@H]1C[C@@H](C2)C(=O)NCC1=CN=NN1)C(=O)OCc1cc(cc(c1)Cl)Cl with IC50=0.326297 microM | | Descriptor: | (3,5-dichlorophenyl)methyl (3aR,5r,6aS)-5-{[(1H-1,2,3-triazol-5-yl)methyl]carbamoyl}hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
8TZC
 
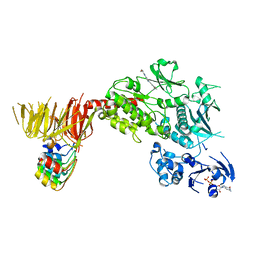 | | Structure of C-terminal LRRK2 bound to MLi-2 (G2019S mutant) | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, E11 DARPin, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanz-Murillo, M, Villagran-Suarez, A, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
7G64
 
 | | Crystal Structure of rat Autotaxin in complex with [(3aS,6aS)-5-[5-cyclopropyl-6-(2,2,2-trifluoroethoxy)pyridine-3-carbonyl]-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-(1H-benzotriazol-5-yl)methanone, i.e. SMILES N1(C[C@@H]2[C@@H](C1)CN(C2)C(=O)c1cc(c(nc1)OCC(F)(F)F)C1CC1)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.0111778 microM | | Descriptor: | ACETATE ION, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6J
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[(2-tert-butyl-4-chloro-5-methylphenoxy)methyl]pyridine, i.e. SMILES c1c(nccc1)COc1cc(c(cc1C(C)(C)C)Cl)C with IC50=1.98688 microM | | Descriptor: | 2-[(2-tert-butyl-4-chloro-5-methylphenoxy)methyl]pyridine, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
