2FEL
 
 | | 3-carboxy-cis,cis-muconate lactonizing enzyme from Agrobacterium radiobacter S2 | | Descriptor: | 3-carboxy-cis,cis-muconate lactonizing enzyme, CHLORIDE ION, SULFATE ION | | Authors: | Lehtio, L, Goldman, A. | | Deposit date: | 2005-12-16 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the 3-carboxy-cis,cis-muconate lactonizing enzyme from the protocatechuate degradative pathway of Agrobacterium radiobacter S2.
Febs J., 273, 2006
|
|
2EWL
 
 | |
2FEN
 
 | | 3-carboxy-cis,cis-muconate lactonizing enzyme from Agrobacterium radiobacter S2 | | Descriptor: | 3-carboxy-cis,cis-muconate lactonizing enzyme, CHLORIDE ION, SULFATE ION | | Authors: | Lehtio, L, Goldman, A. | | Deposit date: | 2005-12-16 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of the 3-carboxy-cis,cis-muconate lactonizing enzyme from the protocatechuate degradative pathway of Agrobacterium radiobacter S2.
Febs J., 273, 2006
|
|
2F57
 
 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
2FDE
 
 | | Wild type HIV protease bound with GW0385 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-3-[(1,3-BENZODIOXOL-5-YLSULFONYL)(ISOBUTYL)AMINO]-2-HYDROXY-1-{4-[(2-METHYL-1,3-THIAZOL-4-YL)METHOXY]BENZYL}PROPYL]CARBAMATE, Gag-Pol polyprotein, POTASSIUM ION | | Authors: | Xu, R.X. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: The discovery of GW0385.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FE9
 
 | |
3PWD
 
 | | Crystal structure of maize CK2 in complex with NBC (Z1) | | Descriptor: | 8-hydroxy-4-methyl-9-nitro-2H-benzo[g]chromen-2-one, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Mazzorana, M. | | Deposit date: | 2010-12-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features underlying the selectivity of the kinase inhibitors NBC and dNBC: role of a nitro group that discriminates between CK2 and DYRK1A
Cell.Mol.Life Sci., 69, 2012
|
|
3PS2
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | Descriptor: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
3QGW
 
 | | Crystal Structure of ITK kinase bound to an inhibitor | | Descriptor: | 3-[(8-phenylthieno[2,3-h]quinazolin-2-yl)amino]benzenesulfonamide, N-(6-oxo-1,6-dihydro-3,4'-bipyridin-5-yl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Cheetham, G.M.T. | | Deposit date: | 2011-01-25 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-activity relationship of 3-aminopyrid-2-ones as potent and selective interleukin-2 inducible T-cell kinase (Itk) inhibitors
J.Med.Chem., 54, 2011
|
|
3QAH
 
 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
3QGY
 
 | | Crystal structure of ITK inhibitor complex | | Descriptor: | 3-[(8-phenylthieno[2,3-h]quinazolin-2-yl)amino]benzenesulfonamide, N-{5-[2-(methylamino)pyrimidin-4-yl]-2-oxo-1,2-dihydropyridin-3-yl}-4-(piperidin-1-yl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Cheetham, G.M.T. | | Deposit date: | 2011-01-25 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-activity relationship of 3-aminopyrid-2-ones as potent and selective interleukin-2 inducible T-cell kinase (Itk) inhibitors
J.Med.Chem., 54, 2011
|
|
3QN7
 
 | | Potent and selective bicyclic peptide inhibitor (UK18) of human urokinase-type plasminogen activator(uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, Bicyclic peptide inhibitor, Urokinase-type plasminogen activator | | Authors: | Angelini, A, Cendron, L, Touati, J, Winter, G, Zanotti, G, Heinis, C. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bicyclic peptide inhibitor reveals large contact interface with a protease target
Acs Chem.Biol., 7, 2012
|
|
3RMF
 
 | | CDK2 in complex with inhibitor RC-2-33 | | Descriptor: | 4-{[4-amino-5-(naphthalen-2-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3R8A
 
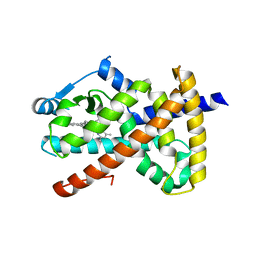 | |
3QTX
 
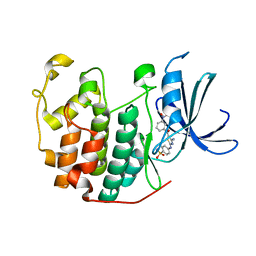 | | CDK2 in complex with inhibitor RC-2-35 | | Descriptor: | 4-{[4-amino-5-(3-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3QXP
 
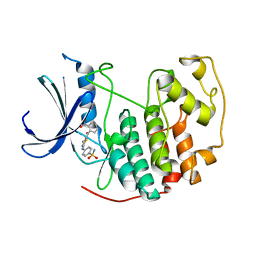 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-02 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RJW
 
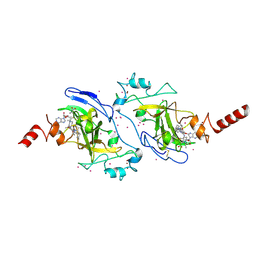 | | Crystal structure of histone lysine methyltransferase g9a with an inhibitor | | Descriptor: | 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Wasney, G.A, Tempel, W, Liu, F, Barsyte, D, Allali-Hassani, A, Chen, X, Chau, I, Hajian, T, Senisterra, G, Chavda, N, Arora, K, Siarheyeva, A, Kireev, D.B, Herold, J.M, Bochkarev, A, Bountra, C, Weigelt, J, Edwards, A.M, Frye, S.V, Arrowsmith, C.H, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells.
Nat.Chem.Biol., 7, 2011
|
|
3PRO
 
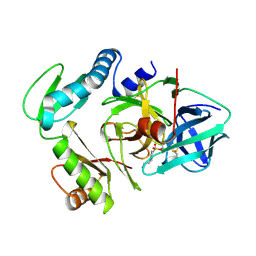 | | ALPHA-LYTIC PROTEASE COMPLEXED WITH C-TERMINAL TRUNCATED PRO REGION | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ALPHA-LYTIC PROTEASE | | Authors: | Sauter, N.K, Mau, T, Rader, S.D, Agard, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of alpha-lytic protease complexed with its pro region.
Nat.Struct.Biol., 5, 1998
|
|
3PLL
 
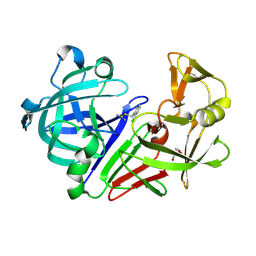 | | Endothiapepsin in complex with a fragment | | Descriptor: | 2-chlorobenzyl carbamimidothioate, Endothiapepsin, GLYCEROL | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3D78
 
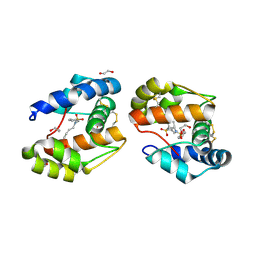 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3PMY
 
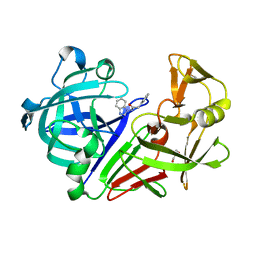 | | Endothiapepsin in complex with a fragment | | Descriptor: | Endothiapepsin, GLYCEROL, N-(1H-benzimidazol-1-yl)-2-phenylacetamide | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3QTU
 
 | | CDK2 in complex with inhibitor RC-2-132 | | Descriptor: | 4-{[4-amino-5-(4-sulfamoylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3QTZ
 
 | | CDK2 in complex with inhibitor RC-2-36 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(3-fluorobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3D77
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 4.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3QU0
 
 | | CDK2 in complex with inhibitor RC-2-38 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(pyridin-3-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
