7WWW
 
 | |
9C7S
 
 | |
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
5JYB
 
 | | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nocek, B, Jedrzejczak, R, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases
To Be Published
|
|
6Y74
 
 | |
5T6G
 
 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
9HD3
 
 | | Crystal structure of human TRF1 TRFH domain in complex with compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 4-ethoxynaphthalene-1-sulfonic acid, CALCIUM ION, ... | | Authors: | Casale, G, Le Bihan, Y.-V, Van Montfort, R.L.M, Guettler, S. | | Deposit date: | 2024-11-11 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of first-in-class inhibitors of the TRF1-TIN2 protein-protein interaction by fragment screening
To Be Published
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
9I08
 
 | | The Paulinella chromatophore transit peptide part2 (crTPpart2) from ArgC | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Klimenko, V, Reiners, J, Applegate, V, Reimann, K, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2025-01-14 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Paulinella chromatophore transit peptide part2 adopts a structural fold similar to the gamma-glutamyl-cyclotransferase fold.
Plant Physiol., 199, 2025
|
|
7XCZ
 
 | |
6E7P
 
 | | cryo-EM structure of human TRPML1 with PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
9QUD
 
 | | Cu(II)-bound de novo protein scaffold TFD-EH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Wagner Egea, P, Delhommel, F, Mustafa, G, Leiss-Maier, F, Klimper, L, Badmann, T, Heider, A, Wille, I.C, Groll, M, Sattler, M, Zeymer, C. | | Deposit date: | 2025-04-10 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modular protein scaffold architecture and AI-guided sequence optimization facilitate de novo metalloenzyme engineering.
Structure, 2025
|
|
6A86
 
 | | Pholiota squarrosa lectin | | Descriptor: | (3R)-butane-1,3-diol, lectin | | Authors: | Yamasaki, K, Yamasaki, T, Kubota, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific recognition of core fucosylation in N-glycans by Pholiota squarrosa lectin (PhoSL).
Glycobiology, 29, 2019
|
|
6YG2
 
 | | Crystal structure of MKK7 (MAP2K7) in complex with ibrutnib, with covalent and allosteric binding modes | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3~{R})-3-[4-azanyl-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, ... | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
4QYX
 
 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6EAS
 
 | | Co-crystal of pseudokinase DRIK1 (drought responsive inactive kinase 1) bound to ENMD-2076 | | Descriptor: | 6-(4-methylpiperazin-1-yl)-N-(5-methyl-1H-pyrazol-3-yl)-2-[(E)-2-phenylethenyl]pyrimidin-4-amine, GLYCEROL, drought responsive inactive kinase 1 | | Authors: | Aquino, B, Counago, R.M, Fala, A.M, Massirer, K.B, Elkins, J.M, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal of pseudokinase DRIK1 with ENMD-2076
To be Published
|
|
9FF3
 
 | | The structure of delta-ScoC, a global regulator protein from Geobacillus kaustophilus T-1 | | Descriptor: | HTH-type transcriptional regulator Hpr | | Authors: | Hadad, N, Shulami, S, Pomyalov, S, Shoham, Y, Shoham, G. | | Deposit date: | 2024-05-22 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The structure of delta-ScoC, a global regulator protein from Geobacillus kaustophilus T-1
To Be Published
|
|
4Z86
 
 | |
7BA0
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 63 | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,28-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.2.2.113,17.04,9]nonacosa-1(26),13(29),14,16,24,27-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7U56
 
 | |
7XDA
 
 | |
8OMI
 
 | | Crystal structure of the inositol hexakisphosphate kinase EhIP6KA M85 variant in complex with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schuetz, A, Aguirre, T, Fiedler, D. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An unconventional gatekeeper mutation sensitizes inositol hexakisphosphate kinases to an allosteric inhibitor.
Elife, 12, 2023
|
|
7X4X
 
 | | BTB domain of KEAP1 in complex with MEF | | Descriptor: | 4-ethoxy-4-oxobutanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Qu, L.Z. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Characterization of the modification of Kelch-like ECH-associated protein 1 by different fumarates.
Biochem.Biophys.Res.Commun., 605, 2022
|
|
6JYQ
 
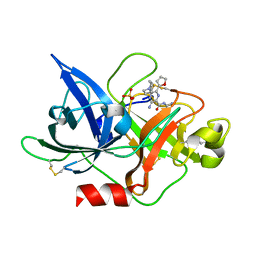 | | Crystal structure of uPA_H99Y in complex with 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | H99Y-6F-HMA-pH7
To Be Published
|
|
