7TBB
 
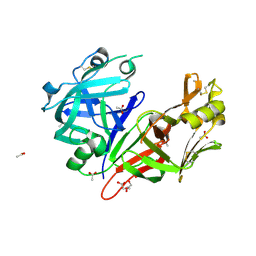 | | Crystal structure of Plasmepsin X from Plasmodium falciparum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Christensen, J.B, Hodder, A.N, Dietrich, M.H, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
7WDB
 
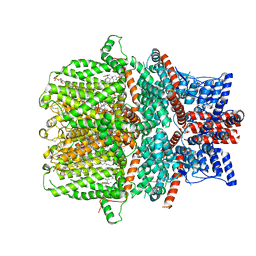 | | Human TRPC5 channel in complex with riluzole | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, CALCIUM ION, ... | | Authors: | Chen, L, Wei, M, Yang, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-04-13 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural identification of riluzole-binding site on human TRPC5.
Cell Discov, 8, 2022
|
|
7TAM
 
 | |
7TAY
 
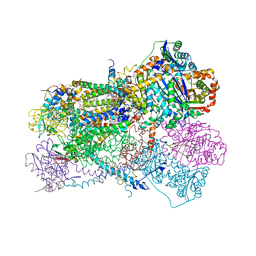 | | Bos Taurus Mitochondrial BC1 in complex with Pyramoxadone | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-3-anilino-5-methyl-5-(6-phenoxypyridin-3-yl)-1,3-oxazolidine-2,4-dione, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conformation Switch of Rieske ISP subunit is revealed by the Crystal Structure of Bacterial Cytochrome bc1 in Complex with Azoxystrobin
to be published
|
|
7TAZ
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor | | Descriptor: | 2-[4-bromo-3-(3-chloro-5-cyanophenoxy)-2-fluorophenyl]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase p51, Reverse transcriptase/ribonuclease H | | Authors: | Snyder, A.A, Risener, C.J, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2021-12-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor
To Be Published
|
|
7QNP
 
 | | Designed Armadillo repeat protein N(A4)M4C(AII) co-crystallized with hen egg white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Designed Armadillo Repeat Protein N(A4)M4C(AII), ... | | Authors: | Michel, E, Mittl, P.R.E, Plueckthun, A. | | Deposit date: | 2021-12-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Improved Repeat Protein Stability by Combined Consensus and Computational Protein Design.
Biochemistry, 62, 2023
|
|
7WD3
 
 | | Cryo-EM structure of Drg1 hexamer treated with ATP and benzo-diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WDJ
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid
To Be Published
|
|
7WD6
 
 | | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXANOIC ACID, ... | | Authors: | Sugiyama, S, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid
To Be Published
|
|
7WDH
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDI
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDE
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7QMT
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 3) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMS
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 2) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMU
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 4) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMV
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 5) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMW
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 6) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMX
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 7) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN0
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 10) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN2
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 12) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WCQ
 
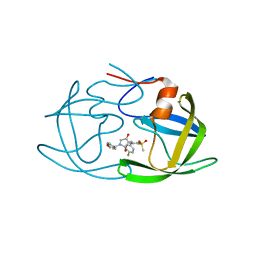 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7TAH
 
 | |
7TAJ
 
 | |
