4HFU
 
 | | Crystal structure of Fab 8M2 in complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 8M2 heavy chain, Fab 8M2 light chain, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1OYT
 
 | | COMPLEX OF RECOMBINANT HUMAN THROMBIN WITH A DESIGNED FLUORINATED INHIBITOR | | Descriptor: | (3ASR,4RS,8ASR,8BRS)-4-(2-(4-FLUOROBENZYL)-1,3-DIOXODEACAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL)BENZAMIDINE, CALCIUM ION, Hirudin IIB, ... | | Authors: | Banner, D.W, Olsen, J.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Fluorine Scan of Thrombin Inhibitors to Map the Fluorophilicity/Fluorophobicity of an Enzyme Active Site: Evidence for CF...C=O Interactions.
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1PN4
 
 | | Crystal structure of 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 complexed with (3R)-hydroxydecanoyl-CoA. | | Descriptor: | 1,2-ETHANEDIOL, 3R-HYDROXYDECANOYL-COENZYME A, Peroxisomal hydratase-dehydrogenase-epimerase | | Authors: | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2003-06-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
1SJ4
 
 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
1S31
 
 | | Crystal Structure Analysis of the human Tub protein (isoform a) spanning residues 289 through 561 | | Descriptor: | TRIETHYLENE GLYCOL, tubby isoform a | | Authors: | Boutboul, S, Carroll, K.J, Basdevant, A, Gomez, C, Nandrot, E, Clement, K, Shapiro, L, Abitbol, M. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | A novel human obesity and sensory deficit syndrome resulting from a mutation in the TUB gene
To be Published
|
|
4HG4
 
 | | Crystal structure of Fab 2G1 in complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G1 heavy chain, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-10-06 | | Release date: | 2013-02-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2O01
 
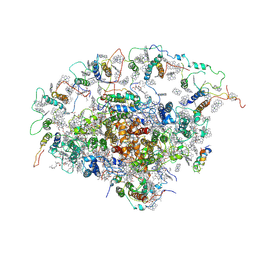 | | The Structure of a plant photosystem I supercomplex at 3.4 Angstrom resolution | | Descriptor: | AT3g54890, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Amunts, A, Drory, O, Nelson, N. | | Deposit date: | 2006-11-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structure of a plant photosystem I supercomplex at 3.4 A resolution.
Nature, 447, 2007
|
|
2PVL
 
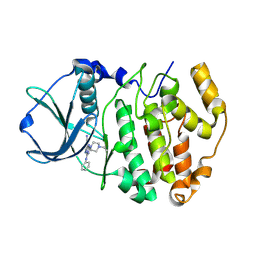 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-ETHYLPIPERAZIN-1-YL)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4HZP
 
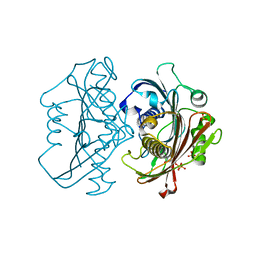 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, CHLORIDE ION, COENZYME A, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
2JT7
 
 | |
2GAR
 
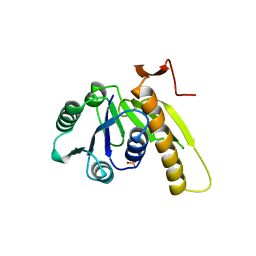 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
1Z19
 
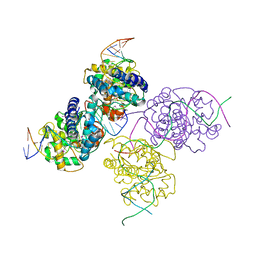 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | Descriptor: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
2NEO
 
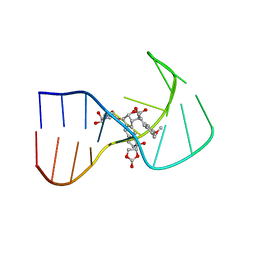 | | SOLUTION NMR STRUCTURE OF A TWO-BASE DNA BULGE COMPLEXED WITH AN ENEDIYNE CLEAVING ANALOG, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*AP*TP*GP*CP*PGE*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3'), SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Stassinopoulos, A, Ji, J, Gao, X, Goldberg, I.H. | | Deposit date: | 1997-06-19 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-base DNA bulge complexed with an enediyne cleaving analog.
Science, 272, 1996
|
|
2FXU
 
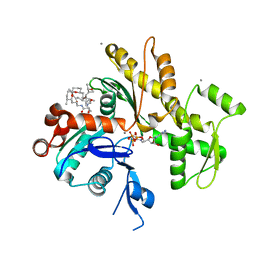 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
1SQX
 
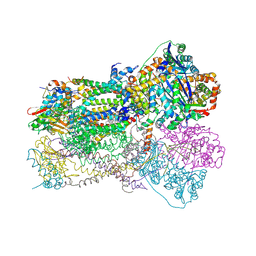 | | Crystal Structure Analysis of Bovine Bc1 with Stigmatellin A | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
1J4J
 
 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
1H6I
 
 | |
2J2I
 
 | | Crystal Structure of the humab PIM1 in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1, SULFATE ION | | Authors: | Debreczeni, J.E, Bullock, A.N, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2H5M
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus complexed with acetyl-CoA. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Cort, J.R, Ramelot, T.A, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins
J.Struct.Funct.Genom., 9, 2008
|
|
2NOV
 
 | | Breakage-reunion domain of S.pneumoniae topo IV: crystal structure of a gram-positive quinolone target | | Descriptor: | DNA topoisomerase 4 subunit A | | Authors: | Laponogov, I, Veselkov, D.A, Sohi, M.K, Pan, X.S, Achari, A, Yang, C, Ferrara, J.D, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
PLoS ONE, 2, 2007
|
|
2V4E
 
 | | A non-cytotoxic DsRed variant for whole-cell labeling | | Descriptor: | RED FLUORESCENT PROTEIN DRFP583 | | Authors: | Strack, R.L, Strongin, D.E, Bhattacharyya, D, Tao, W, Berman, A, Broxmeyer, H.E, Keenan, R.J, Glick, B.S. | | Deposit date: | 2008-09-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Noncytotoxic Dsred Variant for Whole-Cell Labeling.
Nat.Methods, 5, 2008
|
|
2H9R
 
 | | Docking and dimerization domain (D/D) of the regulatory subunit of the Type II-alpha cAMP-dependent protein kinase A associated with a Peptide derived from an A-kinase anchoring protein (AKAP) | | Descriptor: | 22-mer from A-kinase anchor protein 5, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
Embo J., 20, 2001
|
|
2DRN
 
 | | Docking and dimerization domain (D/D) of the Type II-alpha regulatory subunity of protein kinase A (PKA) in complex with a peptide from an A-kinase anchoring protein | | Descriptor: | 24-residues peptide from an a-kinase anchoring protein, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 2006-06-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
Embo J., 20, 2001
|
|
2I5W
 
 | |
1CPF
 
 | | A CATION BINDING MOTIF STABILIZES THE COMPOUND I RADICAL OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miller, M.A, Han, G.W, Kraut, J. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
