5UE6
 
 | | Structure of nitrite reductase AniA from Neisseria gonorrhoeae, space group I4122 | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Hamza, A, Williamson, Z.A, Reed, R.W, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA.
Antimicrob. Agents Chemother., 61, 2017
|
|
8G6I
 
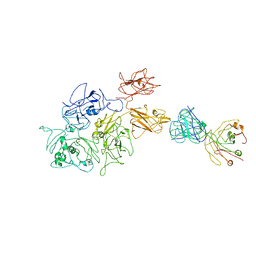 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
5TB7
 
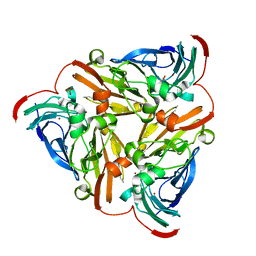 | | Structure of nitrite reductase AniA from Neisseria gonorrhoeae, space group P212121 | | Descriptor: | COPPER (II) ION, Nitrite reductase, PHOSPHATE ION | | Authors: | Hamza, A, Williamson, Z.A, Reed, R.W, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2016-09-11 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA.
Antimicrob. Agents Chemother., 61, 2017
|
|
5ZLL
 
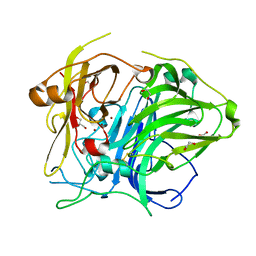 | | Mutation in the trinuclear site of CotA-laccase: H493C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5ZLJ
 
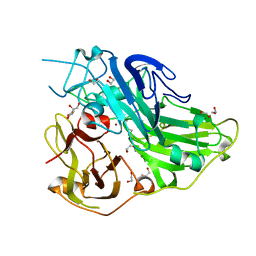 | | Crystal structure of CotA native enzyme, PH8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
1RZQ
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH5.0 | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1RZP
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH6.2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1SJM
 
 | | Nitrite bound copper containing nitrite reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Tocheva, E.I, Rosell, F.I, Mauk, A.G, Murphy, M.E.P. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side-on copper-nitrosyl coordination by nitrite reductase.
Science, 304, 2004
|
|
5I6O
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 20.70 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
8P9V
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H/D268N from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, PEROXIDE ION, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
8P9U
 
 | | Crystal Structure of Two-Domain Laccase mutant M199A/D268N from Streptomyces griseoflavus | | Descriptor: | CALCIUM ION, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
5I6M
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 7.59 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I6P
 
 | |
3KW8
 
 | | Two-domain laccase from Streptomyces coelicolor at 2.3 A resolution | | Descriptor: | COPPER (II) ION, FE (III) ION, Putative copper oxidase, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Duskova, J, Stepankova, A, Hasek, J, Ostergaard, L.H, Ostergaard, P.R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 A
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3J2Q
 
 | | Model of membrane-bound factor VIII organized in 2D crystals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Stoilova-Mcphie, S, Lynch, G.C, Ludtke, S, Pettitt, B.M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (15 Å) | | Cite: | Domain organization of membrane-bound factor VIII.
Biopolymers, 99, 2013
|
|
8QGF
 
 | |
3KW7
 
 | | Crystal structure of LacB from Trametes sp. AH28-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ge, H.H, Teng, M.K, Niu, L.W. | | Deposit date: | 2009-12-01 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure of native laccase B from Trametes sp. AH28-2
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6KLG
 
 | | Crystal Structure of the Zea Mays laccase 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6KNG
 
 | | CryoEM map and model of Nitrite Reductase at pH 8.1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
7KWO
 
 | | rFVIIIFc-VWF-XTEN (BIVV001) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Fuller, J.R, Batchelor, J.D. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy.
Blood, 137, 2021
|
|
7KVE
 
 | |
7KXY
 
 | |
7KVF
 
 | |
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
7BDN
 
 | | Structure of the Streptomyces coelicolor small laccase - cubic crystal form | | Descriptor: | COPPER (II) ION, Putative copper oxidase | | Authors: | Zovo, K, Majumdar, S, Lukk, T. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of the Methionine Axial Ligand of the T1 Copper for the Fungal-like Phenylalanine Ligand (M298F) Causes Local Structural Perturbations that Lead to Thermal Instability and Reduced Catalytic Efficiency of the Small Laccase from Streptomyces coelicolor A3(2).
Acs Omega, 7, 2022
|
|
