4XX3
 
 | |
9NIU
 
 | | Co-crystal structure of FABP7 in complex with PFO3TDA | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, brain, ... | | Authors: | Yang, D, Liu, J, Zeng, H, Dong, A, Arrowsmith, C.H, Edwards, A.M, Peng, H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-02-26 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of FABP7 in complex with PFO3TDA
To be published
|
|
7QDQ
 
 | | Crystal Structure of HDM2 in complex with Caylin-1 | | Descriptor: | CHLORIDE ION, Caylin-1, DIMETHYL SULFOXIDE, ... | | Authors: | Finke, A.D, Walti, M.A, Marsh, M.E, Orts, J. | | Deposit date: | 2021-11-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidation of a nutlin-derivative-HDM2 complex structure at the interaction site by NMR molecular replacement: A straightforward derivation
J Magn Reson Open, 10-11, 2022
|
|
7DQJ
 
 | | E. coli GyrB ATPase domain in complex with 3,4-Dihydroxyacetophenone | | Descriptor: | 1-[3,4-bis(oxidanyl)phenyl]ethanone, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
9ED7
 
 | | Active state of mTOR on membrane | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTP-binding protein Rheb, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2024-11-16 | | Release date: | 2025-09-10 | | Last modified: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for mTORC1 activation on the lysosomal membrane.
Nature, 647, 2025
|
|
6PL2
 
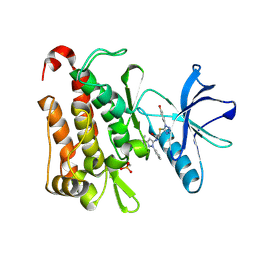 | | TRK-A IN COMPLEX WITH LIGAND 1a | | Descriptor: | High affinity nerve growth factor receptor, N-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-2-{[1-(4-hydroxyphenyl)-1H-tetrazol-5-yl]sulfanyl}acetamide | | Authors: | Subramanian, G. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Type 2 inhibitor leads of human tropomyosin receptor kinase (hTrkA).
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4QEO
 
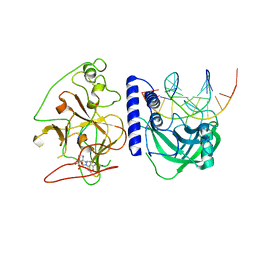 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
9ED8
 
 | | Intermediate state of mTOR on membrane | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTP-binding protein Rheb, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2024-11-16 | | Release date: | 2025-09-10 | | Last modified: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis for mTORC1 activation on the lysosomal membrane.
Nature, 647, 2025
|
|
6TCA
 
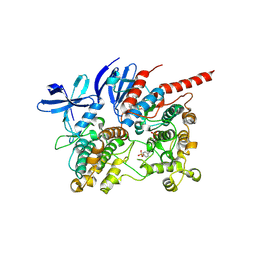 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5GJF
 
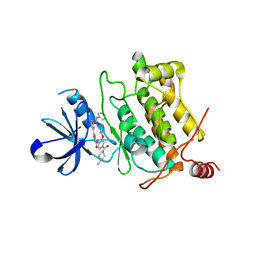 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 3 | | Descriptor: | N-(2-isopropoxy-4-(4-methylpiperazine-1-carbonyl)phenyl)-2-(3-(3-phenylureido)phenyl)thiazole-4-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
5CSW
 
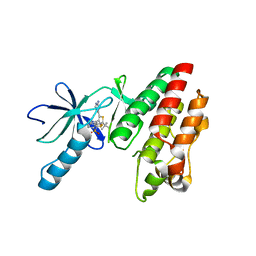 | | B-RAF in complex with Dabrafenib | | Descriptor: | CHLORIDE ION, Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Bader, G, Stadtmuller, H, Steurer, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A Novel RAF Kinase Inhibitor with DFG-Out-Binding Mode: High Efficacy in BRAF-Mutant Tumor Xenograft Models in the Absence of Normal Tissue Hyperproliferation.
Mol.Cancer Ther., 15, 2016
|
|
6V9E
 
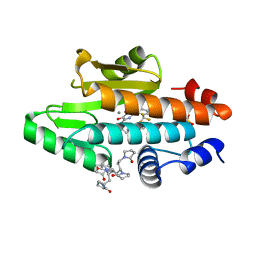 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5T99
 
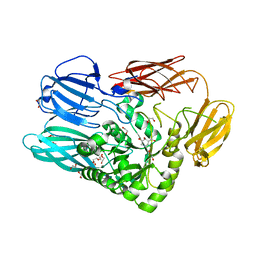 | |
7SNV
 
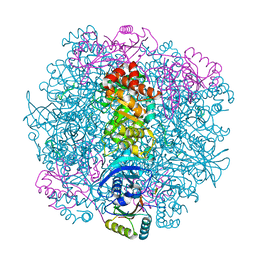 | | H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex | | Descriptor: | Carboxysome shell carbonic anhydrase, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Blikstad, C, Dugan, E, Laughlin, T.G, Liu, M, Shoemaker, S, Remis, J, Savage, D.F. | | Deposit date: | 2021-10-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ZX0
 
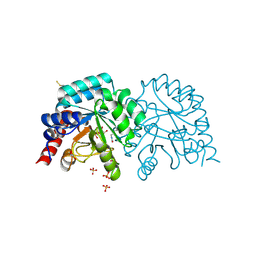 | | OMPD-domain of human UMPS in complex with the substrate OMP at 1.25 Angstroms resolution | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
4QIP
 
 | |
7QWU
 
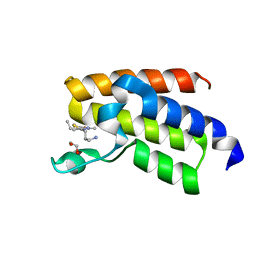 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 44 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-[2-(3-azanylpropyl)-1,3-thiazol-4-yl]-4-ethyl-2-methyl-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7YN3
 
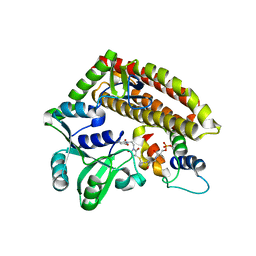 | | Crystal structure of CcbD complex with CcbZ carrier protein domain | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, CcbD, ... | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of CcbD complex with carrier protein
To Be Published
|
|
7QWF
 
 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethyl-2-methyl-5-[2-[2-(methylamino)ethyl]-1,3-thiazol-4-yl]-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7QWY
 
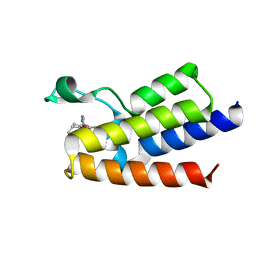 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 61 | | Descriptor: | 1-[4-ethyl-2-methyl-5-[2-(piperazin-1-ylmethyl)-1,3-thiazol-4-yl]-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
6ZWY
 
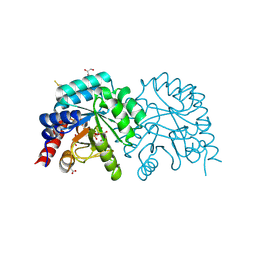 | |
8VCS
 
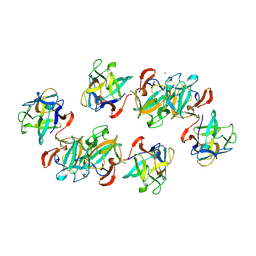 | | Crystal structure of the oligomeric rMcL-1 in complex with lactose | | Descriptor: | CALCIUM ION, Galactose-binding lectin, beta-D-galactopyranose, ... | | Authors: | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
7QXL
 
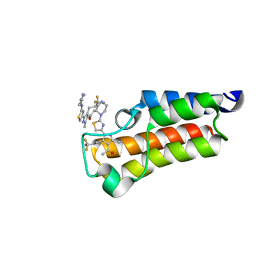 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 77 | | Descriptor: | 1-[2-methyl-5-(2-piperazin-1-yl-1,3-thiazol-4-yl)-4-[3,3,3-tris(fluoranyl)propyl]-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7QX2
 
 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 63 | | Descriptor: | 1-[5-[2-[(4-azanylpiperidin-1-yl)methyl]-1,3-thiazol-4-yl]-4-ethyl-2-methyl-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
