8HRF
 
 | |
6Y5Q
 
 | | human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
8R2K
 
 | | Human Carbonic Anhydrase II in complex with 4-((2-oxo-5-(3,4,5-trimethoxyphenyl)-2,5-dihydrofuran-3-yl)amino)benzenesulfonamide | | Descriptor: | 4-[[(2~{R})-5-oxidanylidene-2-(2,4,5-trimethoxyphenyl)-2~{H}-furan-4-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthetic Approaches to Novel Human Carbonic Anhydrase Isoform Inhibitors Based on Pyrrol-2-one Moiety.
J.Med.Chem., 67, 2024
|
|
8TRQ
 
 | | T cell recognition of citrullinated vimentin peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A07 TCR alpha chain, A07 TCR beta chain, ... | | Authors: | Loh, T.J, Lim, J.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
9G6Z
 
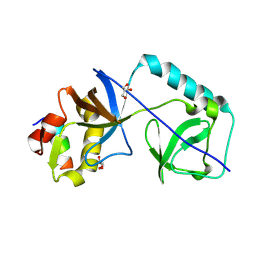 | |
4Y9H
 
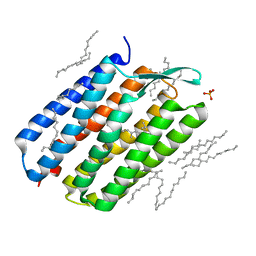 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
8HW1
 
 | | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | | Descriptor: | (1S)-4-[(1E,3Z,5E,7E,9E,11E,13E,15E,17E)-3-(hydroxymethyl)-7,12,16-trimethyl-18-[(1R,4S)-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, CHLOROPHYLL A, ... | | Authors: | Kosugi, M, Kawasaki, M, Shibata, Y, Moriya, T, Adachi, N, Senda, T. | | Deposit date: | 2022-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Uphill energy transfer mechanism for photosynthesis in an Antarctic alga.
Nat Commun, 14, 2023
|
|
6YG7
 
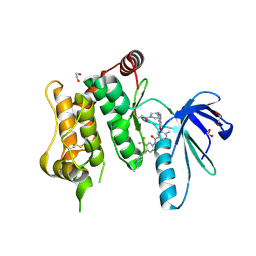 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor SB1-G-23 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{R})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
8R0Y
 
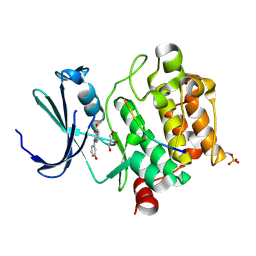 | |
6PVS
 
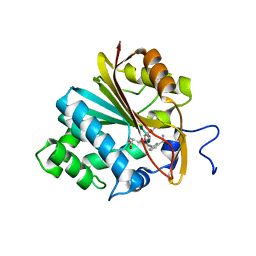 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | Descriptor: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6N93
 
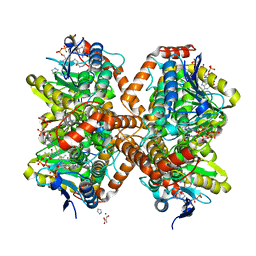 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2E)-2-(hydroxyimino)propanoic acid, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
6SHH
 
 | |
6DQP
 
 | |
7AW1
 
 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
5MCC
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 1.11 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6W9U
 
 | | Structure of human MAIT A-F7 TCR in complex with patient MR1-R9H-Ac-6-FP | | Descriptor: | 1-[[6-(1-$l^{1}-oxidanylethyl)-4-$l^{3}-oxidanylidene-2,3,6,8~{a}-tetrahydropteridin-2-yl]-$l^{2}-azanyl]ethanone, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1 .
Sci Immunol, 5, 2020
|
|
6GNJ
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
9KPM
 
 | | Crystal structure of KRAS-G12C in complex with compound 16 (JAB-16) | | Descriptor: | 7-[2-azanyl-3,5-bis(chloranyl)-6-fluoranyl-phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | Deposit date: | 2024-11-23 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
