6SRB
 
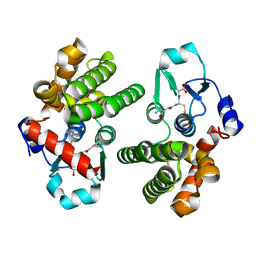 | | Crystal structure of glutathione transferase Omega 3C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSL
 
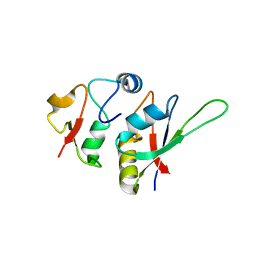 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-09-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
6SRL
 
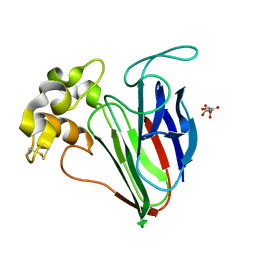 | |
6SRT
 
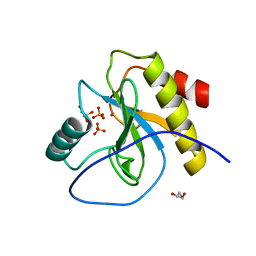 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
6SRP
 
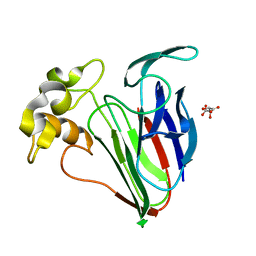 | |
7A8T
 
 | | Crystal structure of sarcomeric protein FATZ-1 (mini-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
6SSE
 
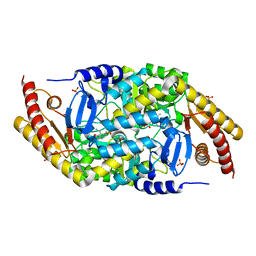 | | Transaminase with PMP bound | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ForI-PMP, SULFATE ION | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SSS
 
 | | Crystal structure of Human Microsomal Glutathione S-Transferase 2 | | Descriptor: | 1-(8Z-hexadecenoyl)-sn-glycerol, GLYCEROL, Microsomal glutathione S-transferase 2, ... | | Authors: | Thulasingam, M, Nji, E, Haeggstrom, J.Z. | | Deposit date: | 2019-09-09 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structures of human MGST2 reveal synchronized conformational changes regulating catalysis.
Nat Commun, 12, 2021
|
|
5LU2
 
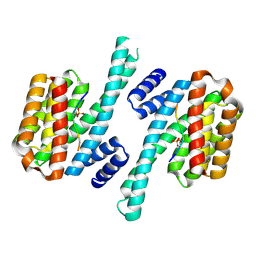 | | Human 14-3-3 sigma complexed with long HSPB6 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Heat shock protein beta-6 | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
7A3G
 
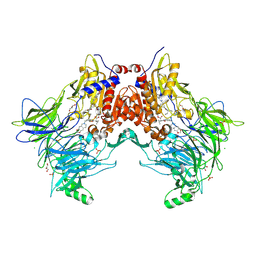 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, 91 | | Descriptor: | 1-[3-(7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-6-ylmethyl)phenyl]-3,3-diethyl-azetidine-2,4-dione, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
7A3J
 
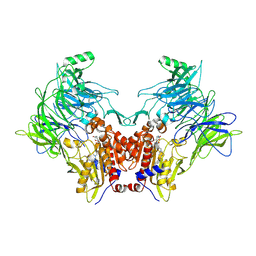 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, A272 | | Descriptor: | 2-ethyl-2-methanoyl-~{N}-[3-[[4-(naphthalen-1-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, CHLORIDE ION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 2022
|
|
6SUI
 
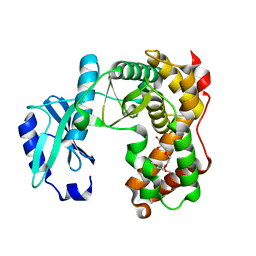 | | AMICOUMACIN KINASE AMIN | | Descriptor: | PENTAETHYLENE GLYCOL, Phosphotransferase enzyme family protein | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
7A3L
 
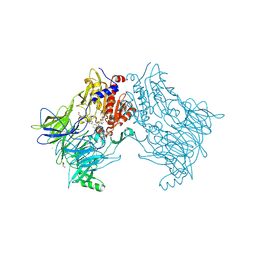 | |
6SU4
 
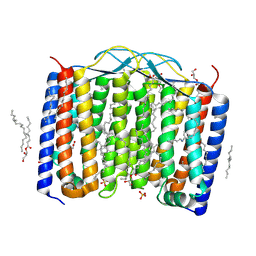 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5LGR
 
 | | Crystal structure of mouse CARM1 in complex with ligand P1C3u | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6SUH
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone, a tolcapone analogue | | Descriptor: | 3-O-methyltolcapone, Transthyretin | | Authors: | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | Deposit date: | 2019-09-14 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
5LVV
 
 | | Human OGT in complex with UDP and fused substrate peptide (Tab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Raimi, O. | | Deposit date: | 2016-09-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
5LW9
 
 | | Crystal structure of human JARID1B in complex with S40563a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-[4-[3,5-bis(chloranyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Le Bihan, Y.V, Szykowska, A, Johansson, C, Gileadi, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Huber, K. | | Deposit date: | 2016-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human JARID1B in complex with S40563a
to be published
|
|
7A3I
 
 | |
7ABM
 
 | | X-ray structure of phosphorylated Barrier-to-autointegration factor (BAF) | | Descriptor: | Barrier-to-autointegration factor, CESIUM ION | | Authors: | Zinn-Justin, S, Marcelot, A, Le Du, M.H, Ropars, V. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
5LX2
 
 | |
9G98
 
 | | Joint neutron and x-ray structure of alginate lyase PsAlg7C soaked with pentamannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Wilkens, C, Meilleur, F, Morth, J.P. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-30 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
5LXV
 
 | |
