1NM9
 
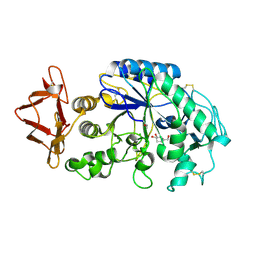 | | Crystal structure of recombinant human salivary amylase mutant W58A | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J, Thomas, L.M. | | Deposit date: | 2003-01-09 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
1N2K
 
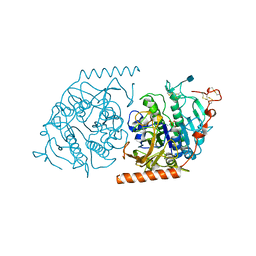 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
3VGP
 
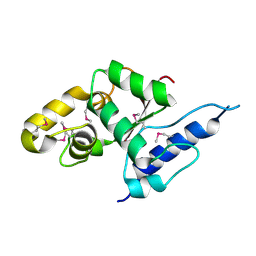 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
3WAI
 
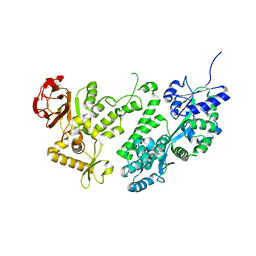 | |
1WZ7
 
 | | Crystal structure of enhancer of rudimentary homologue (ERH) | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Arai, R, Kukimoto-Niino, M, Uda-Tochio, H, Morita, S, Uchikubo-Kamo, T, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-26 | | Release date: | 2005-05-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an enhancer of rudimentary homolog (ERH) at 2.1 Angstroms resolution.
Protein Sci., 14, 2005
|
|
1Q4N
 
 | |
1QY2
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOPROPYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1QY1
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1IPC
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
2HZY
 
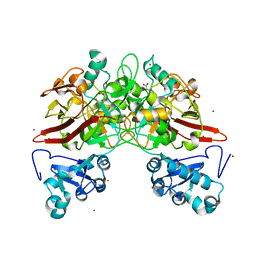 | |
1E33
 
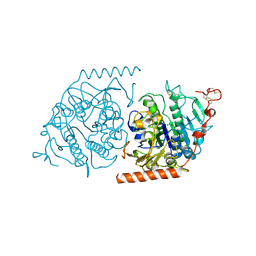 | | Crystal structure of an Arylsulfatase A mutant P426L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-06 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy.
J. Biol. Chem., 277, 2002
|
|
1GM6
 
 | | 3-D STRUCTURE OF A SALIVARY LIPOCALIN FROM BOAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Spinelli, S, Vincent, F, Pelosi, P, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Boar Salivary Lipocalin. Three-Dimensional X-Ray Structure and Androsterol/Androstenone Docking Simulations.
Eur.J.Biochem., 269, 2002
|
|
1JXJ
 
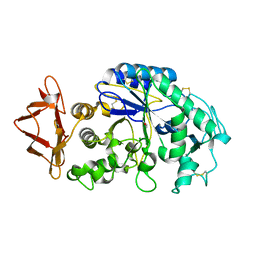 | | Role of mobile loop in the mechanism of human salivary amylase | | Descriptor: | Alpha-amylase, salivary, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Wang, Z, Mishra, P.J, Thomas, L.M. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
1SIR
 
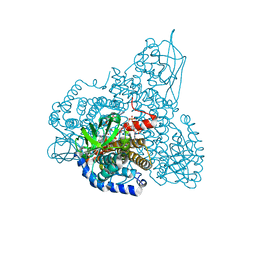 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, S-4-NITROBUTYRYL-COA | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S.L, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
1SIQ
 
 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
2PN0
 
 | | Prokaryotic transcription elongation factor GreA/GreB from Nitrosomonas europaea | | Descriptor: | Prokaryotic transcription elongation factor GreA/GreB | | Authors: | Osipiuk, J, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of prokaryotic transcription elongation factor GreA/GreB from Nitrosomonas europaea.
To be Published
|
|
1ZEO
 
 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
5L2P
 
 | | Structure of arylesterase | | Descriptor: | Arylesterase | | Authors: | Lee, H.B, Park, Y.J. | | Deposit date: | 2016-08-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of arylesterase
To Be Published
|
|
8RS9
 
 | | p97 (VCP) double mutant - F266A F539A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) double mutant - F266A F539A
To Be Published
|
|
8RSB
 
 | | p97 (VCP) mutant - F539A ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) mutant - F539A ADP state
To Be Published
|
|
8R0E
 
 | | p97 (VCP) mutant - F266A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | p97 (VCP) mutant - F266A
To Be Published
|
|
8RSC
 
 | | p97 (VCP) mutant - F539A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | p97 (VCP) mutant - F539A
To Be Published
|
|
5DC3
 
 | | Complex of yeast 80S ribosome with non-modified eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-23 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of Hypusine-Containing Translation Factor eIF5A Bound to a Rotated Eukaryotic Ribosome.
J.Mol.Biol., 428, 2016
|
|
8PQX
 
 | | p97 (VCP) mutant - F539A state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | p97 (VCP) mutant - F539A state III
To Be Published
|
|
5NVM
 
 | |
