4XP4
 
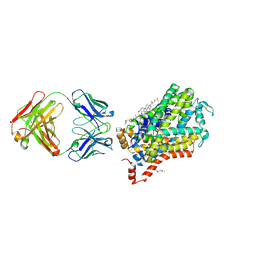 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XP1
 
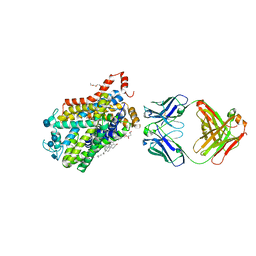 | | X-ray structure of Drosophila dopamine transporter bound to neurotransmitter dopamine | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XP5
 
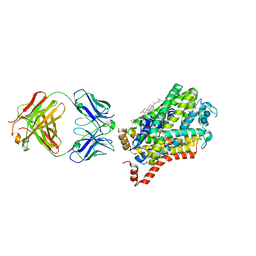 | | X-ray structure of Drosophila dopamine transporter bound to cocaine analogue-RTI55 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XPB
 
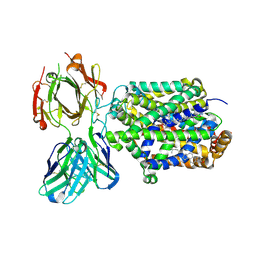 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) bound to cocaine | | Descriptor: | Antibody fragment heavy chain-protein, 9D5-heavy chain, Antibody fragment light chain-protein, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XP6
 
 | | X-ray structure of Drosophila dopamine transporter bound to psychostimulant methamphetamine | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody fragment heavy chain-protein, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XPH
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) bound to 3,4dichlorophenethylamine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-(3,4-dichlorophenyl)ethanamine, Antibody fragment heavy chain, ... | | Authors: | Penmatsa, A, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XP9
 
 | | X-ray structure of Drosophila dopamine transporter bound to psychostimulant D-amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XPA
 
 | | X-ray structure of Drosophila dopamine transporter bound to 3,4dichlorophenethylamine | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4BWZ
 
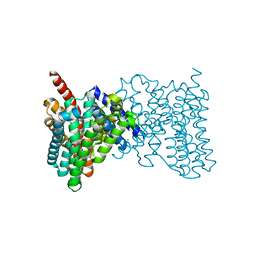 | |
7DIX
 
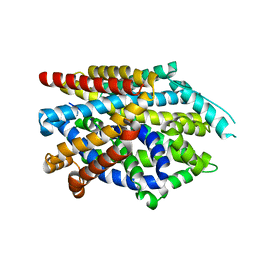 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
5BZ2
 
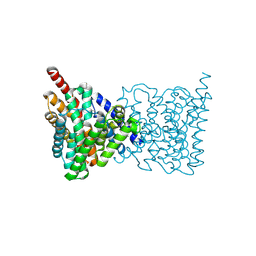 | |
1JTW
 
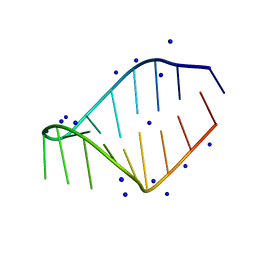 | |
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
2M7X
 
 | | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2 | | Descriptor: | Na(+)/H(+) antiporter | | Authors: | Ullah, A, Kemp, G, Lee, B, Alves, C, Young, H, Sykes, B.D, Fliegel, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2.
J.Biol.Chem., 288, 2013
|
|
2KRG
 
 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
4D0A
 
 | |
4CZB
 
 | | Structure of the sodium proton antiporter MjNhaP1 from Methanocaldococcus jannaschii at pH 8. | | Descriptor: | NA(+)/H(+) ANTIPORTER 1, POTASSIUM ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Woehlert, D, Paulino, C, Kapotova, E, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 3D Em Map of the Sodium Proton Antiporter Mjnhap1 from Methanocaldococcus Jannaschii
Elife, 3, 2014
|
|
6V1Q
 
 | | Two-pore channel 3 | | Descriptor: | SODIUM ION, Two pore channel 3 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Resting state structure of the hyperdepolarization activated two-pore channel 3.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QS5
 
 | |
3QS6
 
 | |
3QS4
 
 | |
6L85
 
 | | The sodium-dependent phosphate transporter | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Sun, Y.-J. | | Deposit date: | 2019-11-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of the sodium-dependent phosphate transporter reveals insights into human solute carrier SLC20.
Sci Adv, 6, 2020
|
|
7LW1
 
 | | Human phosphofructokinase-1 liver type bound to activator NA-11 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B. | | Deposit date: | 2021-02-27 | | Release date: | 2022-01-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Selective activation of PFKL suppresses the phagocytic oxidative burst.
Cell, 184, 2021
|
|
4O5Z
 
 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), SODIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6L9S
 
 | | Crystal structure of Na-dithionite reduced auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, COPPER (I) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
