8DOJ
 
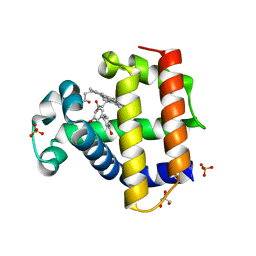 | | Dehaloperoxidase B in complex with 3,3'-Biphenol | | Descriptor: | (1P)-[1,1'-biphenyl]-3,3'-diol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
3MNN
 
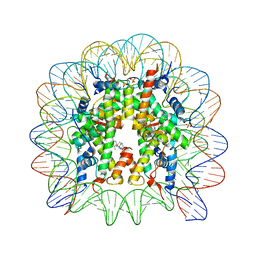 | | A Ruthenium Antitumour Agent Forms Specific Histone Protein Adducts in the Nucleosome Core | | Descriptor: | 1,3,5-triaza-7-phosphatricyclo[3.3.1.1~3,7~]decane, 1-methyl-4-(1-methylethyl)benzene, DNA (145-MER), ... | | Authors: | Ong, M.S, Davey, C.A. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ruthenium antimetastasis agent forms specific histone protein adducts in the nucleosome core
Chemistry, 17, 2011
|
|
4MPX
 
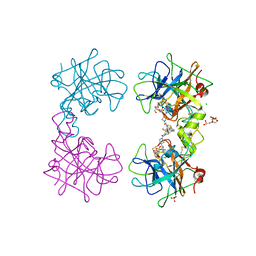 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzothiene-4,2-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Target-directed self-assembly of homodimeric drugs
To be Published
|
|
7REV
 
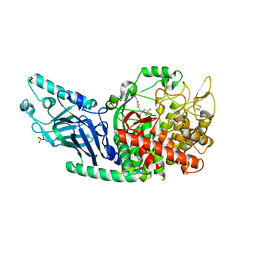 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7YHW
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6QTP
 
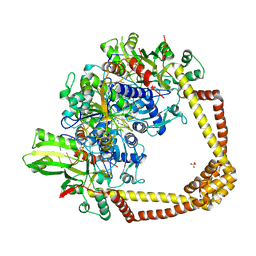 | | 2.37A structure of gepotidacin with S.aureus DNA gyrase and uncleaved DNA | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*AP*GP*CP*TP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-02-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Acs Infect Dis., 5, 2019
|
|
9BHZ
 
 | | Structure of FbsH, an NRPS adenylation domain in the fimsbactin biosynthetic pathway bound to Salicyl-AMS | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydroxybenzoate-AMP ligase, 5'-O-[(2-hydroxybenzoyl)sulfamoyl]adenosine | | Authors: | Ahmed, S.F, Gulick, A.M. | | Deposit date: | 2024-04-22 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Expanding the Substrate Selectivity of the Fimsbactin Biosynthetic Adenylation Domain, FbsH.
Acs Chem.Biol., 19, 2024
|
|
9BHY
 
 | | Structure of FbsH, an NRPS adenylation domain in the fimsbactin biosynthetic pathway bound to 2,3-dihydroxybenzoic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase | | Authors: | Ahmed, S.F, Gulick, A.M. | | Deposit date: | 2024-04-22 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the Substrate Selectivity of the Fimsbactin Biosynthetic Adenylation Domain, FbsH.
Acs Chem.Biol., 19, 2024
|
|
9BCV
 
 | | Cyclase domain of GC-A bound to ANP | | Descriptor: | Atrial natriuretic peptide receptor 1, Fab fragment, MAGNESIUM ION, ... | | Authors: | Liu, S, Huang, X. | | Deposit date: | 2024-04-09 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture and activation of single-pass transmembrane receptor guanylyl cyclase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8AM6
 
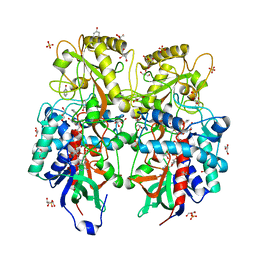 | |
8XM5
 
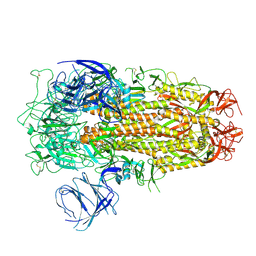 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
6QXA
 
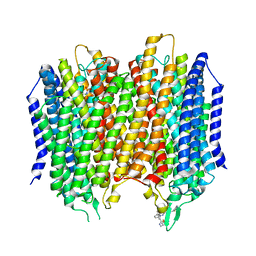 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
8S39
 
 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with isophthalic acid and NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
6REY
 
 | | Human 20S-PA200 Proteasome Complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Toste Rego, A, da Fonseca, P.C.A. | | Deposit date: | 2019-04-12 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes.
Mol.Cell, 76, 2019
|
|
8AJ1
 
 | | SARS-CoV-2 Mpro in Complex with RK-107 | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(phenylcarbamoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2024-09-11 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
7R0L
 
 | | Structure of the FK1 domain of the FKBP51 G64S variant in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Meyners, C, Hausch, F. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding pocket stabilization by high-throughput screening of yeast display libraries.
Front Mol Biosci, 9, 2022
|
|
8PNH
 
 | | Chorismate mutase | | Descriptor: | 3-PHENYLPYRUVIC ACID, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Bifunctional cyclohexadienyl dehydratase/chorismate mutase from Janthinobacterium sp. HH01, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chorismate mutase
To Be Published
|
|
6EPK
 
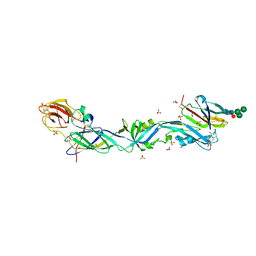 | | CRYSTAL STRUCTURE OF THE PRECURSOR MEMBRANE PROTEIN-ENVELOPE PROTEIN HETERODIMER FROM THE YELLOW FEVER VIRUS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, GLYCEROL, ... | | Authors: | Rey, F.A, Duquerroy, S, Crampon, E, Barba-Spaeth, G. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New insight into flavivirus maturation from structure/function studies of the yellow fever virus envelope protein complex
Mbio, 2023
|
|
9BA8
 
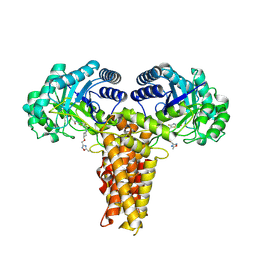 | | O-GlcNAcase (OGA) inhibitor complex for the Treatment of Alzheimer's Disease | | Descriptor: | N-[4-fluoro-5-({(2S,4S)-2-methyl-4-[(5-methyl-1,2,4-oxadiazol-3-yl)methoxy]piperidin-1-yl}methyl)-1,3-thiazol-2-yl]acetamide, Protein O-GlcNAcase | | Authors: | Hendle, J, Romero, R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery and clinical translation of ceperognastat, an O-GlcNAcase (OGA) inhibitor, for the treatment of Alzheimer's disease.
Alzheimers Dement (N Y), 10, 2024
|
|
8B8K
 
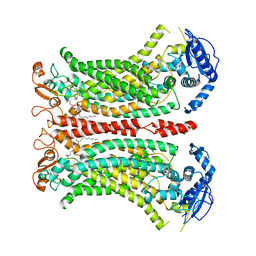 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
9F7F
 
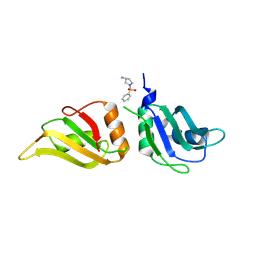 | | UP1 in complex with Z1491353358 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
9F63
 
 | |
5J1Y
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 1-(pyrrolidin-1-yl)-3-(tetrahydrofuran-3-yl)propan-1-one at 1.81A resolution | | Descriptor: | 3-[(3S)-oxolan-3-yl]-1-(pyrrolidin-1-yl)propan-1-one, EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
9F4P
 
 | | UP1 in complex with Z1217960891 | | Descriptor: | (3S)-3-(3-fluorophenoxy)-1-methylpyrrolidin-2-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
9F4O
 
 | | UP1 in complex with Z991506900 | | Descriptor: | (3~{R})-3-methyl-1-(6-methylpyridin-2-yl)piperazine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
