6UM8
 
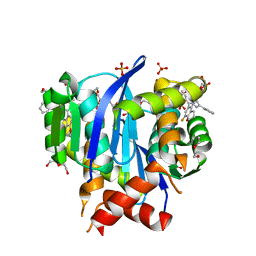 | | HIV Integrase in complex with Compound-14 | | Descriptor: | (2S)-tert-butoxy[7-(8-fluoro-5-methyl-3,4-dihydro-2H-1-benzopyran-6-yl)-5-methyl-2-phenylpyrazolo[1,5-a]pyrimidin-6-yl]acetic acid, DI(HYDROXYETHYL)ETHER, Integrase, ... | | Authors: | Khan, J.A, Kish, K. | | Deposit date: | 2019-10-09 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of Novel Pyrazolopyrimidines as Potent and Orally Bioavailable Allosteric HIV-1 Integrase Inhibitors.
J.Med.Chem., 63, 2020
|
|
7SVO
 
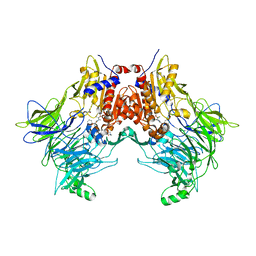 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
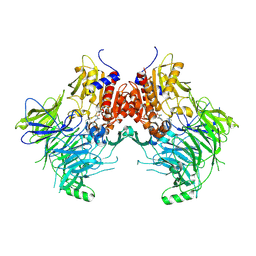 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
4HNI
 
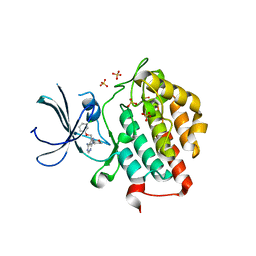 | | crystal structure of ck1e in complex with PF4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
9NTB
 
 | | Crystal structure of human HDAC2 in complex with TNG260 | | Descriptor: | (R)-N-(4-amino-4'-fluoro-[1,1'-biphenyl]-3-yl)-4-(S-methylsulfonimidoyl)benzamide, 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | McMillan, B.J, Whittington, D.A. | | Deposit date: | 2025-03-18 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TNG260 Is a Small-Molecule CoREST Inhibitor That Sensitizes STK11-Mutant Tumors to Anti-PD-1 Immunotherapy.
Cancer Res., 85, 2025
|
|
6H1L
 
 | | Structure of the BM3 heme domain in complex with tioconazole | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
5Y0F
 
 | |
6F3J
 
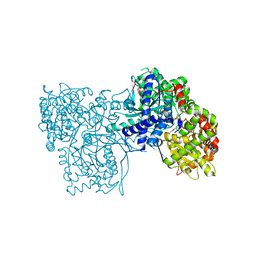 | | The crystal structure of Glycogen Phosphorylase in complex with 10a | | Descriptor: | 4-[4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]phenyl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6UDX
 
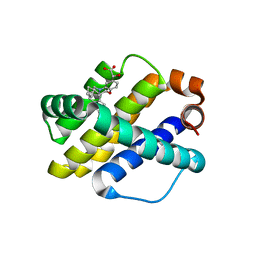 | | X-ray co-crystal structure of compound 7 with Mcl-1 | | Descriptor: | (2R)-[(3S)-6'-chloro-5-(cyclobutylmethyl)-3',4,4',5-tetrahydro-2H,2'H-spiro[1,5-benzoxazepine-3,1'-naphthalen]-7-yl](hydroxy)acetic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6F1X
 
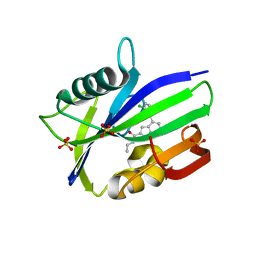 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
8HE8
 
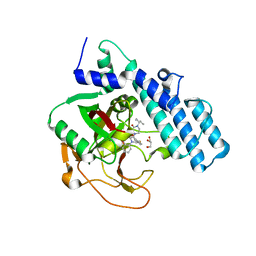 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
6OLP
 
 | | Full length HIV-1 Env AMC011 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K, Cottrell, C.A. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
6F3R
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10c | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(9~{H}-fluoren-2-yl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
8TD0
 
 | | Structure of PYCR1 complexed with 5-oxo-7a-phenyl-hexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid | | Descriptor: | (3R,4S,7aR)-5-oxo-7a-phenylhexahydropyrrolo[2,1-b][1,3]thiazole-3-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
6F3S
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10d | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-phenylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
5Y12
 
 | |
6UUI
 
 | |
5HKX
 
 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
9I8I
 
 | | cryoEM structure of HIV-1 KAKA/G225R mature CA hexamer | | Descriptor: | HIV-1 KAKA/G225R CA hexamer | | Authors: | Zhu, Y, Shen, J, Shen, Y, Xu, J, Zhang, P. | | Deposit date: | 2025-02-05 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for HIV-1 capsid adaption to rescue IP6-packaging deficiency.
Biorxiv, 2025
|
|
1I5V
 
 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
7TXG
 
 | | Structure of the Class II Fructose-1,6-Bisphosphatase from Francisella tularensis with native Mn++ divalent cation and partially occupied product F6P | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Abad-Zapatero, C, Selezneva, A.I, Harding, L.N.M, Movahedzadeh, F. | | Deposit date: | 2022-02-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
8YMH
 
 | | BRD4-BD1 in complex with 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-{[(2R)-2-methyl-4-methylsulfonyl-piperazin-1-yl]methyl}-7-(1-methylpyrazol-3-yl)furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4 | | Authors: | Sasaki, C, Miyaguchi, I, Hagihara, S, Ishizawa, K, Endo, J. | | Deposit date: | 2024-03-09 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Discovery of a potent, orally available furopyridine derivative as a novel selective BET BD1 Inhibitor (Part 1).
To Be Published
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6TVP
 
 | | Structure of Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM | | Descriptor: | Alpha-maltose-1-phosphate synthase, SODIUM ION | | Authors: | Syson, K, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5NX7
 
 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate and 2-fluorogeranyl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
