9HBL
 
 | | SmsC2B2 complex from M. jannaschii (monoclinic form) | | Descriptor: | 1,2-ETHANEDIOL, Iron-sulfur cluster assembly SufBD family protein MJ0034, SULFATE ION, ... | | Authors: | Mechaly, A.E, Dussouchaud, M, Haouz, A, Betton, J.M, Barras, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.
Plos Biol., 23, 2025
|
|
8HTG
 
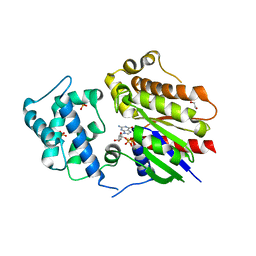 | | Crystal structure of Golf in complex with GTP-gamma S and Mg | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | Authors: | Kang, H, Choi, H.-J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6WLY
 
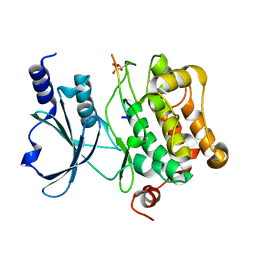 | |
6HPO
 
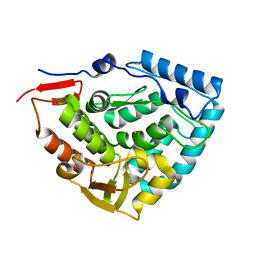 | |
9BRB
 
 | |
8VSY
 
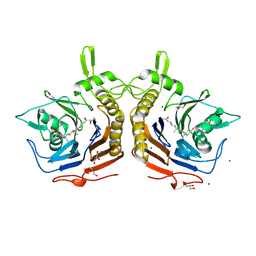 | | Bile salt hydrolase from Arthrobacter citreus with covalent inhibitor AAA-10 bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Bile salt hydrolase, CHLORIDE ION, ... | | Authors: | Dhindwal, P, Ruzzini, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a bile salt hydrolase from the bison microbiome.
J.Biol.Chem., 300, 2024
|
|
5XDL
 
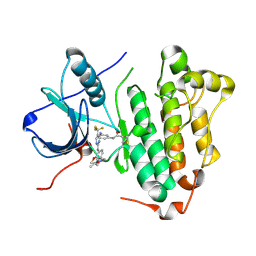 | | Crystal structure of EGFR 696-1022 L858R in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Zhu, S.J, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
8VUW
 
 | | ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-AMINO-ETHANETHIOL, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2024-01-29 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
6YYL
 
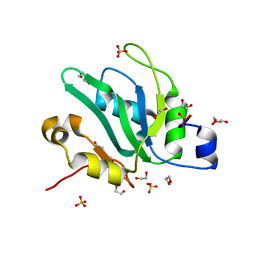 | | Crystal structure of S. pombe Mei2 RRM3 domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Meiosis protein mei2, ... | | Authors: | Graille, M, Hazra, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A scaffold lncRNA shapes the mitosis to meiosis switch.
Nat Commun, 12, 2021
|
|
8VDA
 
 | |
6YZ3
 
 | | PqsR (MvfR) in complex with antagonist 19 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(2-hexyl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, LysR family transcriptional regulator | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
8AY0
 
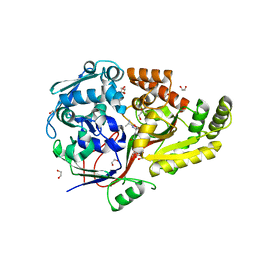 | | Crystal Structure of the peptide binding protein DppE from Bacillus subtilis in complex with murein tripeptide | | Descriptor: | 1,2-ETHANEDIOL, Dipeptide-binding protein DppE, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Hughes, A.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
6YZA
 
 | |
2G3S
 
 | | RNA structure containing GU base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3', MAGNESIUM ION | | Authors: | Jang, S.B, Hung, L.W, Jeong, M.S, Holbrook, E.L, Chen, X, Turner, D.H, Holbrook, S.R. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure at 1.5 angstroms resolution of an RNA octamer duplex containing tandem G.U basepairs
Biophys.J., 90, 2006
|
|
8VD9
 
 | |
8AX2
 
 | |
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
8G9P
 
 | | Tricomplex of RMC-4998, KRAS G12C, and CypA | | Descriptor: | (2S)-2-{(5S)-7-[(2E)-4-(dimethylamino)-4-methylpent-2-enoyl]-1-oxo-2,7-diazaspiro[4.4]nonan-2-yl}-N-[(1P,8S,10R,14S,21M)-22-ethyl-21-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]-3-methylbutanamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS.
Science, 381, 2023
|
|
8PTS
 
 | | human GUK1 in complex with compound AT8001 | | Descriptor: | GLYCEROL, Guanylate kinase, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Zimberger, C, Canard, B, Ferron, F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
6HZ5
 
 | | Structure of McrBC without DNA binding domains (Class 1) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Nirwan, N, Saikrishnan, K, Amunts, A. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC.
Nat Commun, 10, 2019
|
|
8AX0
 
 | |
6WOA
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 2-Diphosphoinositol pentakisphosphate (2-IP7), Mg, and Fluoride ion | | Descriptor: | (1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6OOY
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Tumor necrosis factor, {1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}methanol | | Authors: | Arakaki, T.L, Edwards, T.E, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
7PY4
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dieudonne, T, Abad-Herrera, S, Juknaviciute Laursen, M, Lejeune, M, Stock, C, Slimani, K, Jaxel, C, Lyons, J.A, Montigny, C, Gunther Pomorski, T, Nissen, P, Lenoir, G. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders.
Elife, 11, 2022
|
|
