4BTY
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-(4-methylpiperazin-1-yl)phenylamino]-2-(4-chlorophenyl)-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
3PR2
 
 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
3SLN
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
6T2E
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7B8W
 
 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
5KQ5
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-07-05 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated Protein Kinase (AMPK), for the Potential Treatment of Diabetic Nephropathy.
J.Med.Chem., 59, 2016
|
|
5LH4
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 1 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-(2,2-diphenylethyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5LGO
 
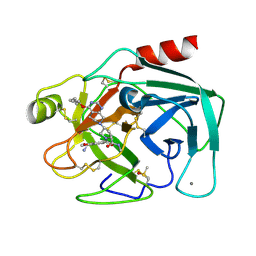 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 15 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-[4-(2-cyclopropylethoxy)-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidin-7-yl]-~{N}-methyl-pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5LH8
 
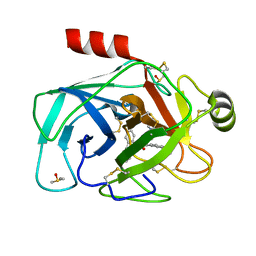 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 8 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-[(1~{S},2~{R})-2-phenylcyclohexyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6T2D
 
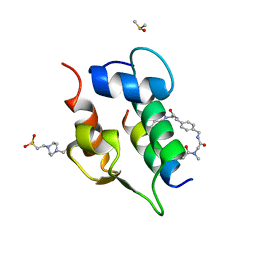 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5RLJ
 
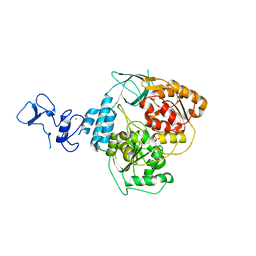 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1407673036 | | Descriptor: | (2S)-2-phenylpropane-1-sulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1741964527 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1101755952 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RL8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53825177 | | Descriptor: | Helicase, N-(2-fluorophenyl)ethanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z285782452 | | Descriptor: | Helicase, N-methyl-2-(methylsulfonyl)aniline, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM9
 
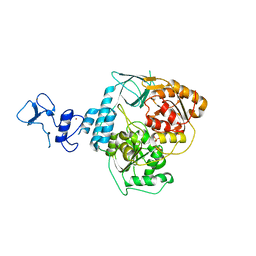 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2856434942 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLI
 
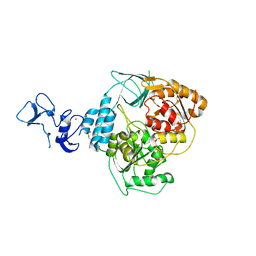 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z45617795 | | Descriptor: | Helicase, N-(2-phenylethyl)methanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLT
 
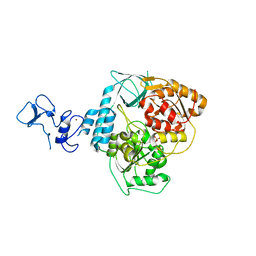 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMB
 
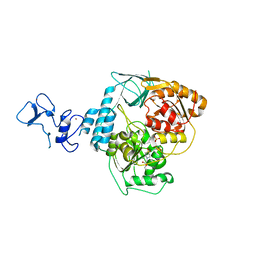 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2856434920 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1509882419 | | Descriptor: | 1-(propan-2-yl)-1H-imidazole-4-sulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z216450634 | | Descriptor: | Helicase, N-cycloheptyl-N-methylmethanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z426041412 | | Descriptor: | Helicase, N-[4-(aminomethyl)phenyl]methanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLP
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z166605480 | | Descriptor: | (1S)-1-(4-fluorophenyl)-N-methylethan-1-amine, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1273312153 | | Descriptor: | Helicase, N-methyl-1H-indole-7-carboxamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
