6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
1W5C
 
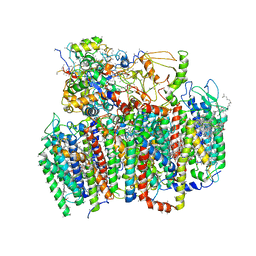 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
7T7Y
 
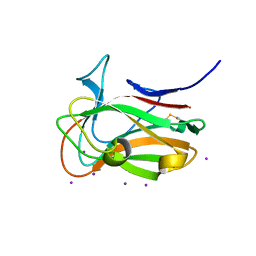 | |
7T7Z
 
 | | The crystal structure of family 8 carbohydrate-binding module from Dictyostelium discoideum | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Endoglucanase, ... | | Authors: | Marcelo, M.V, Campos, B.M, Squina, F.M. | | Deposit date: | 2021-12-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unique properties of a Dictyostelium discoideum carbohydrate-binding module expand our understanding of CBM-ligand interactions.
J.Biol.Chem., 298, 2022
|
|
5UDS
 
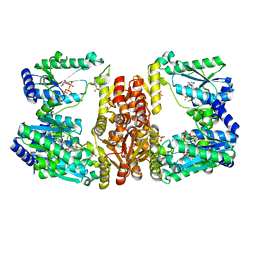 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lactate racemization operon protein LarE, MAGNESIUM ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MLQ
 
 | | Crystal structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7T7G
 
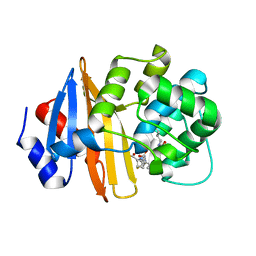 | | Imipenem-OXA-23 2 minute complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7F
 
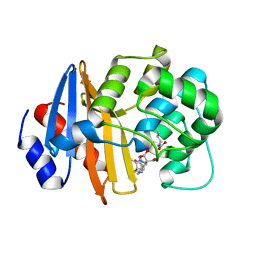 | | MA-1-206-OXA-23 25 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7D
 
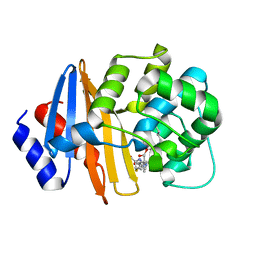 | | MA-1-206-OXA-23 30s complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
1AUL
 
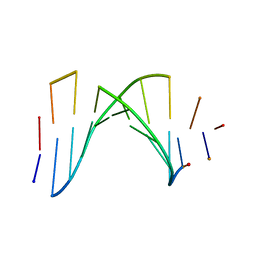 | | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*THXP*GP*AP*TP*TP*AP*TP*CP*TP*G)-3') | | Authors: | Kumar, S, Reed, M.W, Gamper Junior, H.B, Gorn, V.V, Lukhtanov, E.A, Foti, M, West, J, Meyer Junior, R.B, Schweitzer, B.I. | | Deposit date: | 1997-08-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a highly stable DNA duplex conjugated to a minor groove binder.
Nucleic Acids Res., 26, 1998
|
|
7T7E
 
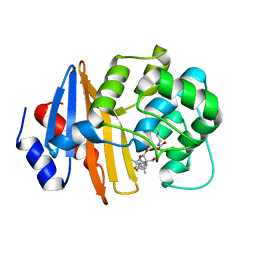 | | MA-1-206-OXA-23 3 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
4P1B
 
 | |
2Y93
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
5FD0
 
 | |
4OPO
 
 | |
1TVX
 
 | |
5TJ2
 
 | |
5TJN
 
 | | Crystal structure of GTB + B trisaccharide (native) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
3NQD
 
 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
7F8W
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
1S3E
 
 | | Crystal structure of MAOB in complex with 6-hydroxy-N-propargyl-1(R)-aminoindan | | Descriptor: | (3R)-3-(PROP-2-YNYLAMINO)INDAN-5-OL, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-13 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1YRJ
 
 | | Crystal Structure of Apramycin bound to a Ribosomal RNA A site oligonucleotide | | Descriptor: | APRAMYCIN, Bacterial 16 S Ribosomal RNA A Site Oligonucleotide, MAGNESIUM ION, ... | | Authors: | Han, Q, Zhao, Q, Fish, S, Simonsen, K.B, Vourloumis, D, Froelich, J.M, Wall, D, Hermann, T. | | Deposit date: | 2005-02-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex.
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
3O9I
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with af61 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
1SGY
 
 | | TYR 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
