5WOL
 
 | | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-hydroxy-tetrahydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii
To Be Published
|
|
5WOS
 
 | |
5WOZ
 
 | |
5WQ7
 
 | |
5WRZ
 
 | | Structure of human PARP1 catalytic domain bound to a phthalazinone inhibitor | | Descriptor: | 7-fluoranyl-4-[[(3R)-pyrrolidin-3-yl]methoxy]-2H-phthalazin-1-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a phthalazinone inhibitor
To Be Published
|
|
6DOH
 
 | |
5WPH
 
 | | Crystal structure of ArsN, N-acetyltransferase with substrate AST from Pseudomonas putida KT2440 | | Descriptor: | (2S)-2-amino-4-[hydroxy(methyl)arsoryl]butanoic acid, Phosphinothricin N-acetyltransferase, SODIUM ION | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-08-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
5WPW
 
 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
5WT3
 
 | |
1EGN
 
 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2000-02-16 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
5WTQ
 
 | | Crystal structure of human proteasome-assembling chaperone PAC4 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Proteasome assembly chaperone 4 | | Authors: | Kurimoto, E, Satoh, T, Ito, Y, Ishihara, E, Tanaka, K, Kato, K. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation
Protein Sci., 26, 2017
|
|
3V7N
 
 | |
5WR8
 
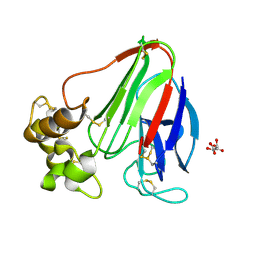 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WRH
 
 | |
3MVK
 
 | | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A | | Descriptor: | GLYCEROL, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Stein, A.J, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A
To be Published
|
|
5WRP
 
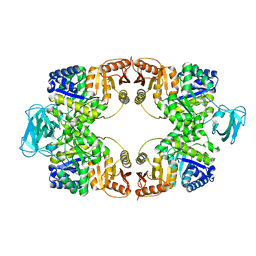 | | T-state crystal structure of pyruvate kinase from Mycobacterium tuberculosis | | Descriptor: | PHOSPHATE ION, Pyruvate kinase | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
3MYF
 
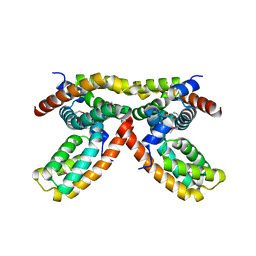 | |
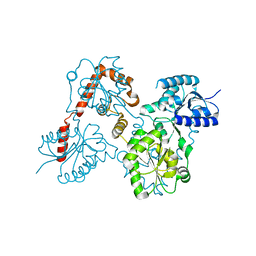
5W8N
 
 | |
5WTP
 
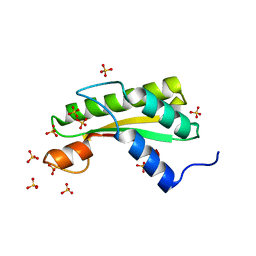 | | Crystal structure of the C-terminal domain of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | OmpA family protein, SULFATE ION | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
6DPI
 
 | |
5WAU
 
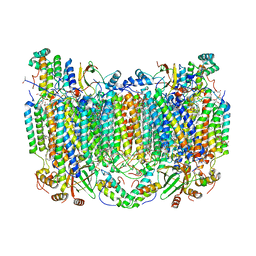 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Synchrotron X-Ray Crystallography at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Fromme, R, Ishigami, I, Yeh, S.Y, Zatsepin, N, Grant, T, Fromme, P, Rousseau, D. | | Deposit date: | 2017-06-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WUY
 
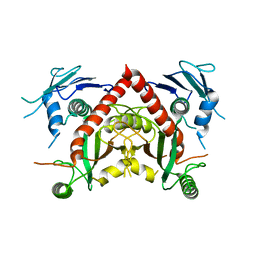 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Iqbal, N, Chaudhary, A, Shukla, K.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To Be Published
|
|
5WVD
 
 | | Structure of Mnk1 in complex with DS12881479 | | Descriptor: | 1-methyl-N-(5-phenyl-1,3-thiazol-2-yl)piperidine-4-carboxamide, MAP kinase interacting serine/threonine kinase 1, SULFATE ION | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2016-12-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel inhibitor stabilizes the inactive conformation of MAPK-interacting kinase 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5W7W
 
 | | Crystal Structure of FHA domain of human APLF | | Descriptor: | Aprataxin and PNK-like factor, FORMIC ACID, SODIUM ION | | Authors: | Pedersen, L.C, Kim, K, London, R.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | Characterization of the APLF FHA-XRCC1 phosphopeptide interaction and its structural and functional implications.
Nucleic Acids Res., 45, 2017
|
|
3MVP
 
 | | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A | | Descriptor: | TetR/AcrR transcriptional regulator | | Authors: | Stein, A.J, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A
To be Published
|
|
