3OP5
 
 | | Human vaccinia-related kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | Authors: | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
3P7F
 
 | | Structure of the human Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Skerra, A, Schiefner, A. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The carbohydrate recognition domain of Langerin reveals high structural similarity with the one of DC-SIGN but an additional, calcium-independent sugar-binding site.
Mol.Immunol., 45, 2008
|
|
3PDK
 
 | | crystal structure of phosphoglucosamine mutase from B. anthracis | | Descriptor: | PHOSPHATE ION, Phosphoglucosamine mutase | | Authors: | Mehra-Chaudhary, R, Mick, J, Tanner, J.J, Henzl, M, Beamer, L.J. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Bacillus anthracis Phosphoglucosamine Mutase, an Enzyme in the Peptidoglycan Biosynthetic Pathway.
J.Bacteriol., 193, 2011
|
|
3PE8
 
 | |
3PJ8
 
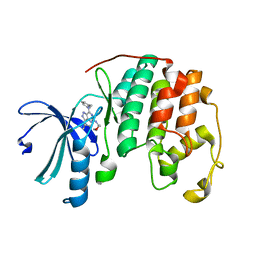 | | Structure of CDK2 in complex with a Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine. | | Descriptor: | (2R)-2-{[7-(benzylamino)-3-(propan-2-yl)-1H-pyrazolo[4,3-d]pyrimidin-5-yl]amino}butan-1-ol, Cell division protein kinase 2 | | Authors: | McNae, I.W, Jorda, R, Havlicek, L, Strnad, M, Voller, J, Walkinshaw, M.D, Krystof, V. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine: Evaluation of a Novel Selective Inhibitor of Cyclin-Dependent Kinases with Antiproliferative Activity.
J.Med.Chem., 54, 2011
|
|
3PDN
 
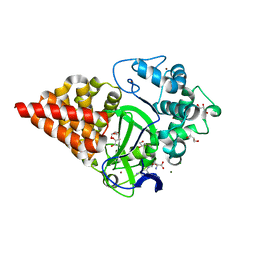 | | Crystal structure of SmyD3 in complex with methyltransferase inhibitor sinefungin | | Descriptor: | GLYCEROL, MAGNESIUM ION, SET and MYND domain-containing protein 3, ... | | Authors: | Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2010-10-23 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Autoinhibition and Posttranslational Activation of Histone Methyltransferase SmyD3.
J.Mol.Biol., 406, 2011
|
|
3PJS
 
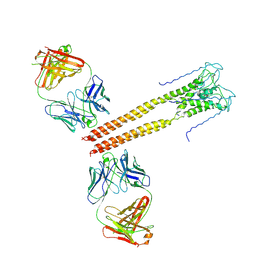 | | Mechanism of Activation Gating in the Full-Length KcsA K+ Channel | | Descriptor: | FAB heavy chain, FAB light chain, Voltage-gated potassium channel | | Authors: | Uysal, S, Cuello, L.G, Kossiakoff, A, Perozo, E. | | Deposit date: | 2010-11-10 | | Release date: | 2011-07-06 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of activation gating in the full-length KcsA K+ channel.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PF5
 
 | |
3PVG
 
 | | Crystal structure of Z. mays CK2 alpha subunit in complex with the inhibitor 4,5,6,7-tetrabromo-1-carboxymethylbenzimidazole (K68) | | Descriptor: | (4,5,6,7-tetrabromo-1H-benzimidazol-1-yl)acetic acid, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2010-12-07 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3PGG
 
 | | Crystal structure of cryptosporidium parvum u6 snrna-associated sm-like protein lsm5 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5. SM domain | | Authors: | Dong, A, Gao, M, Zhao, Y, Lew, J, Wasney, G.A, Kozieradzki, I, Vedadi, M, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Genome-Scale Protein Expression and Structural Biology of Plasmodium Falciparum and Related Apicomplexan Organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3DD6
 
 | | Crystal structure of Rph, an exoribonuclease from Bacillus anthracis at 1.7 A resolution | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Rawlings, A.E, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-06-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The structure of Rph, an exoribonuclease from Bacillus anthracis, at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3PRM
 
 | | Structural analysis of a viral OTU domain protease from the Crimean-Congo Hemorrhagic Fever virus in complex with human ubiquitin | | Descriptor: | Polyubiquitin-B (Fragment), RNA-directed RNA polymerase L | | Authors: | Capodagli, G.C, McKercher, M.A, Baker, E.A, Masters, E.M, Brunzelle, J.S, Pegan, S.D. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin.
J.Virol., 85, 2011
|
|
3DCV
 
 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | Descriptor: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DDW
 
 | | Crystal structure of glycogen phosphorylase complexed with an anthranilimide based inhibitor GSK055 | | Descriptor: | (2S)-{[(3-{[(2-chloro-6-methylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]amino}(phenyl)ethanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2QKV
 
 | |
3DE6
 
 | |
3PQE
 
 | |
2QNX
 
 | | Crystal structure of the complex between the mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and 11-[(decyloxycarbonyl)dithio]-undecanoic acid | | Descriptor: | 11-MERCAPTOUNDECANOIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase 3, O-DECYL HYDROGEN THIOCARBONATE | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
3PQV
 
 | | Cyclase homolog | | Descriptor: | D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tanaka, N, Smith, P, Shuman, S. | | Deposit date: | 2010-11-27 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis.
Rna, 17, 2011
|
|
3PZZ
 
 | |
3D9Y
 
 | |
2QSP
 
 | | Bovine Hemoglobin at pH 5.7 | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Aranda IV, R, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-07-31 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
3DB1
 
 | |
2QUH
 
 | |
3D84
 
 | |
