8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
8QH0
 
 | | Crystal structure of the SARS-CoV-2 RBD with the antibody Cv2.3194 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cv2.3194 Heavy chain, GLYCEROL, ... | | Authors: | Fernandez, I, Rey, F.A. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Broad sarbecovirus neutralization by combined memory B cell antibodies to ancestral SARS-CoV-2.
Iscience, 27, 2024
|
|
8Q3H
 
 | | Capra hircus reactive intermediate deaminase A mutant - A108D | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, GLYCEROL | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Site-directed mutagenesis reveals the interplay between stability, structure, and enzymatic activity in RidA from Capra hircus.
Protein Sci., 33, 2024
|
|
8QPX
 
 | |
8GC2
 
 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8V1V
 
 | |
6NP0
 
 | | Cryo-EM structure of 5HT3A receptor in presence of granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular mechanism of setron-mediated inhibition of full-length 5-HT3Areceptor.
Nat Commun, 10, 2019
|
|
9OMG
 
 | |
6WM3
 
 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WKR
 
 | | PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome | | Descriptor: | DNA (314-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Kasinath, V, Nogales, E, Beck, C, Sauer, P, Poepsel, S, Kosmatka, J, Faini, M, Toso, D, Aebersold, R. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications.
Science, 371, 2021
|
|
8Q87
 
 | | Structure of the G. gallus 80S rotated ribosome in complex with eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Nurullina, L, Jenner, L, Yusupov, M. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of the inactive ribosome complex accumulated in chick embryo cells in cold-stress conditions.
Febs Lett., 598, 2024
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
9OOG
 
 | |
8Q7Z
 
 | | Structure of the G. gallus 80S non-rotated ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Nurullina, L, Jenner, L, Yusupov, M. | | Deposit date: | 2023-08-17 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the inactive ribosome complex accumulated in chick embryo cells in cold-stress conditions.
Febs Lett., 598, 2024
|
|
8W3W
 
 | | Crystal structure of IRAK4 in complex with compound 4 | | Descriptor: | 7-methoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
8W3X
 
 | | Crystal structure of IRAK4 in complex with compound 6 | | Descriptor: | 7-ethoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
8QQB
 
 | | Crystal structure of protein kinase CK2 catalytic subunit in complex with a Dibromo Dihydro Dibenzofuranone derivative | | Descriptor: | (4~{Z})-7,9-bis(bromanyl)-8-oxidanyl-4-(phenylazanylmethylidene)-1,2-dihydrodibenzofuran-3-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Werner, C, Niefind, K, Jose, J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of 7,9-Dibromo-dihydrodibenzofuran as a Potent Casein Kinase 2 (CK2) Inhibitor: Synthesis, Biological Evaluation, and Structural Studies on E- / Z -Isomers.
Acs Pharmacol Transl Sci, 7, 2024
|
|
9Q84
 
 | | Crystal structure of Lotus japonicus CHIP13 extracellular domain in complex with chitooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Andersen, K.R. | | Deposit date: | 2025-02-21 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
9QRS
 
 | | Crystal structure of CERK6 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, J.X.J, Gysel, K, Stougaard, J, Andersen, K.R. | | Deposit date: | 2025-04-04 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
6W9G
 
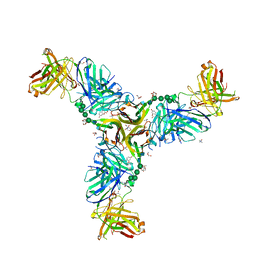 | | Crystal Structure of the Fab fragment of humanized 5c8 antibody containing the fluorescent non-canonical amino acid L-(7-hydroxycoumarin-4-yl)ethylglycine in complex with CD40L at pH 6.8 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5c8* Fab (heavy chain), ... | | Authors: | Henderson, J.N, Simmons, C.R, Mills, J.H. | | Deposit date: | 2020-03-23 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insights into How Protein Environments Tune the Spectroscopic Properties of a Noncanonical Amino Acid Fluorophore.
Biochemistry, 59, 2020
|
|
6VZZ
 
 | |
6X96
 
 | |
6X98
 
 | |
6X97
 
 | |
7Q49
 
 | | Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ZINC ION, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
