1B3Z
 
 | | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOPENTAOSE | | Descriptor: | PROTEIN (XYLANASE), beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Schmidt, A, Kratky, C. | | Deposit date: | 1998-12-15 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant.
Biochemistry, 38, 1999
|
|
9BJ4
 
 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C952 Heavy Chain, C952 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
4RJI
 
 | | Acetolactate synthase from Bacillus subtilis bound to ThDP - crystal form I | | Descriptor: | Acetolactate synthase, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sommer, B, von Moeller, H, Haack, M, Qoura, F, Langner, C, Bourenkov, G, Garbe, D, Brueck, T, Loll, B. | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Detailed Structure-Function Correlations of Bacillus subtilis Acetolactate Synthase.
Chembiochem, 16, 2015
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
2Y2U
 
 | | Nonaged form of Mouse Acetylcholinesterase inhibited by VX-Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETYLCHOLINESTERASE, ... | | Authors: | Akfur, C, Artursson, E, Ekstrom, F. | | Deposit date: | 2010-12-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methylphosphonate Adducts of Acetylcholinesterase Investigated by Time Correlated Single Photon Counting and X-Ray Crystallography
To be Published
|
|
2MD6
 
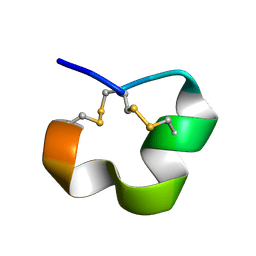 | | NMR SOLUTION STRUCTURE OF ALPHA CONOTOXIN LO1A FROM Conus longurionis | | Descriptor: | ALPHA CONOTOXIN LO1A | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Lebbe, E.K.M, Peigneur, S, D'Souza, L, Tytgat, J. | | Deposit date: | 2013-09-01 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Elucidation of a New alpha-Conotoxin, Lo1a, from Conus longurionis.
J.Biol.Chem., 289, 2014
|
|
3QIZ
 
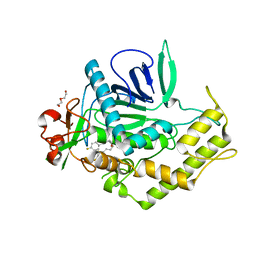 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-2 | | Descriptor: | (2S,4R)-2-(2-{[3-(4-fluoro-3-methylphenyl)propyl](methyl)amino}ethyl)-4-(4-fluorophenyl)-N-hydroxy-4-methoxybutanamide, Botulinum neurotoxin type A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
5O38
 
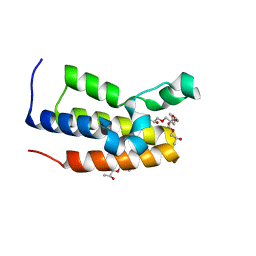 | | Human Brd2(BD2) mutant in free form | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, 3-[(2~{R})-2-oxidanylpropoxy]-2-[[(2~{R})-2-oxidanylpropoxy]methyl]-2-[[(2~{S})-2-oxidanylpropoxy]methyl]propan-1-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
7VVB
 
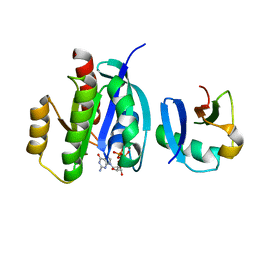 | |
5O3C
 
 | | Human Brd2(BD2) mutant in complex with 9-Me | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
2XYA
 
 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
9BJ3
 
 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1596 Fab Heavy Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
5O3I
 
 | | Human Brd2(BD2) mutant in complex with AL-tBu | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
3FRV
 
 | |
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6V4C
 
 | | Culex quinquefasciatus D7 long form 1- CxD7L1 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Calvo, E, Garboczi, D.N, Martin-Martin, I, Gittis, A.G. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals.
Nat Commun, 11, 2020
|
|
1BD9
 
 | |
7ZM5
 
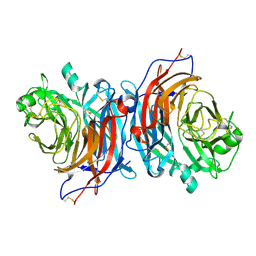 | | Structure of Mossman virus receptor binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Stelfox, A.J, Bowden, T.A, Rissanen, I, Harlos, K. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
7ZM6
 
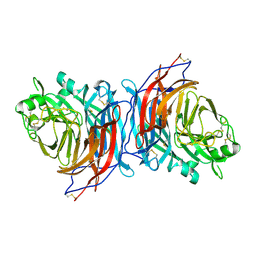 | | Nariva virus receptor binding protein | | Descriptor: | Attachment protein | | Authors: | Stelfox, A.J, Rissanen, I, Rambo, R, Lee, B, Bowden, T.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
2XVZ
 
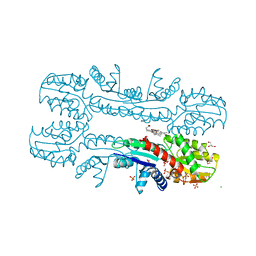 | | Cobalt chelatase CbiK (periplasmatic) from Desulvobrio vulgaris Hildenborough (co-crystallized with cobalt) | | Descriptor: | CHELATASE, PUTATIVE, CHLORIDE ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6ILF
 
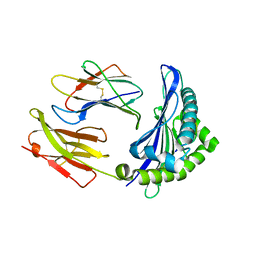 | |
2XKM
 
 | | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | | Descriptor: | CAPSID PROTEIN G8P | | Authors: | Straus, S.K, P Scott, W.R, Schwieters, C.D, Marvin, D.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | FIBER DIFFRACTION (3.3 Å), SOLID-STATE NMR | | Cite: | Consensus Structure of Pf1 Filamentous Bacteriophage from X-Ray Fibre Diffraction and Solid-State NMR.
Eur.Biophys.J., 40, 2011
|
|
5HGG
 
 | | Crystal structure of uPA in complex with a camelid-derived antibody fragment | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Camelid Derived Antibody Fragment, ... | | Authors: | Yung, K.W.Y, Kromann-Hansen, T, Andreasen, P.A, Ngo, J.C.K. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
J.Biol.Chem., 291, 2016
|
|
3QJX
 
 | | Crystal Structure of E. coli Aminopeptidase N in complex with L-Serine | | Descriptor: | Aminopeptidase N, GLYCEROL, MALONATE ION, ... | | Authors: | Addlagatta, A, Gumpena, R, Kishor, C, Ganji, R.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of alpha, beta- and alpha, gamma-Diamino Acid Scaffolds for the Inhibition of M1 Family Aminopeptidases
Chemmedchem, 6, 2011
|
|
5NX2
 
 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
