2JIS
 
 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
2JIT
 
 | | Crystal structure of EGFR kinase domain T790M mutation | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIU
 
 | | Crystal structure of EGFR kinase domain T790M mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIV
 
 | | Crystal structure of EGFR kinase domain T790M mutation in compex with HKI-272 | | Descriptor: | CHLORIDE ION, EPIDERMAL GROWTH FACTOR RECEPTOR, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Li, Y, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIW
 
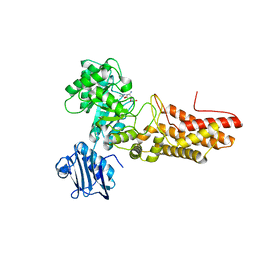 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine | | Descriptor: | N-[(1S,2R,5R,6R)-2-AMINO-5,6-DIHYDROXY-4-(HYDROXYMETHYL)CYCLOHEX-3-EN-1-YL]ACETAMIDE, O-GLCNACASE BT_4395 | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A 1-Acetamido Derivative of 6-Epi-Valienamine: An Inhibitor of a Diverse Group of Beta-N-Acetylglucosaminidases.
Org.Biomol.Chem., 5, 2007
|
|
2JIX
 
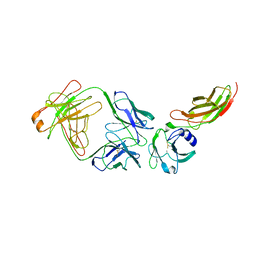 | | Crystal structure of ABT-007 FAB fragment with the soluble domain of EPO receptor | | Descriptor: | ABT-007 FAB FRAGMENT, ERYTHROPOIETIN RECEPTOR | | Authors: | Liu, Z, Stoll, V.S, DeVries, P, Jakob, C.G, Xie, N, Simmer, R.L, Lacy, S.E, Egan, D.A, Harlan, J.E, Lesniewski, R.R, Reilly, E.B. | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-10 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Potent Erythropoietin-Mimicking Human Antibody Interacts Through a Novel Binding Site.
Blood, 110, 2007
|
|
2JIY
 
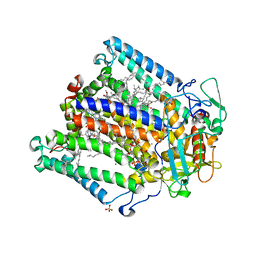 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M149 REPLACED WITH TRP (CHAIN M, AM149W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
2JIZ
 
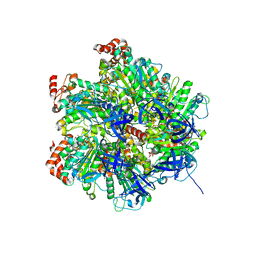 | | The Structure of F1-ATPase inhibited by resveratrol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JJ0
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M248 REPLACED WITH TRP (CHAIN M, AM248W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
2JJ1
 
 | | The Structure of F1-ATPase inhibited by piceatannol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JJ2
 
 | | The Structure of F1-ATPase inhibited by quercetin. | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JJ3
 
 | | Estrogen receptor beta ligand binding domain in complex with a Benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, ESTROGEN RECEPTOR BETA | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Benzopyrans as Selective Estrogen Receptor Beta Agonists (Serbas). Part 4: Functionalization of the Benzopyran A-Ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JJ4
 
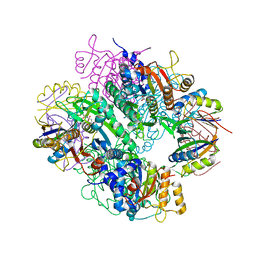 | | The complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, N-ACETYL-L-GLUTAMATE, NITROGEN REGULATORY PROTEIN P-II | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JJ6
 
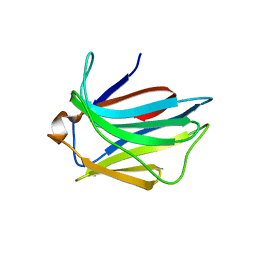 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
2JJ7
 
 | |
2JJ8
 
 | |
2JJ9
 
 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2JJA
 
 | | Crystal structure of GNA with synthetic copper base pair | | Descriptor: | COBALT HEXAMMINE(III), COPPER (II) ION, GNA, ... | | Authors: | Schlegel, M.K, Essen, L.-O, Meggers, E. | | Deposit date: | 2008-03-28 | | Release date: | 2008-07-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Duplex Structure of a Minimal Nucleic Acid.
J.Am.Chem.Soc., 130, 2008
|
|
2JJB
 
 | | Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose | | Descriptor: | 1,2-ETHANEDIOL, CASUARINE, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J, Cardona, F, Parmeggiani, C, Bonaccini, C, Gratteri, P, Sim, L, Rose, D.R, Goti, A. | | Deposit date: | 2008-03-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Total Syntheses of Casuarine and its 6-O-Alpha-Glucoside: Complementary Inhibition Towards Glycoside Hydrolases of the Gh31 and Gh37 Families.
Chemistry, 15, 2009
|
|
2JJC
 
 | | Hsp90 alpha ATPase domain with bound small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA, PYRIMIDIN-2-AMINE | | Authors: | Congreve, M, Chessari, G, Tisi, D, Woodhead, A.J. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recent Developments in Fragment-Based Drug Discovery.
J.Med.Chem., 51, 2008
|
|
2JJD
 
 | | Protein Tyrosine Phosphatase, Receptor Type, E isoform | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE EPSILON | | Authors: | Elkins, J.M, Ugochukwu, E, Alfano, I, Barr, A.J, Bunkoczi, G, King, O.N.F, Filippakopoulos, P, Savitsky, P, Salah, E, Pike, A, Johansson, C, Das, S, Burgess-Brown, N.A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2JJE
 
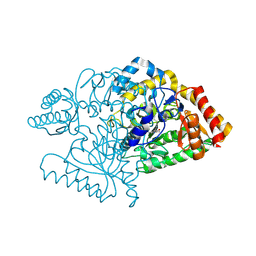 | |
2JJF
 
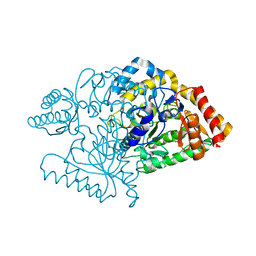 | | N328A mutant of M. tuberculosis Rv3290c | | Descriptor: | L-LYSINE EPSILON AMINOTRANSFERASE | | Authors: | tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
2JJG
 
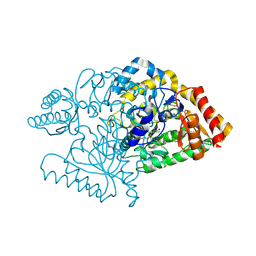 | |
2JJH
 
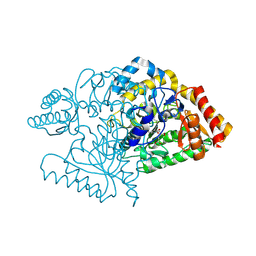 | | E243 mutant of M. tuberculosis Rv3290C | | Descriptor: | 2-OXOGLUTARIC ACID, L-LYSINE EPSILON AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
