4WX7
 
 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6U1I
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, phosphorylated D-cycloserine, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Pederick, J.L, Bruning, J.B, Thompson, A.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
5G4X
 
 | | The crystal structure of the SHANK3 N-terminus | | Descriptor: | 1,2-ETHANEDIOL, SH3 AND MULTIPLE ANKYRIN REPEAT DOMAINS PROTEIN 3 | | Authors: | Zacharchenko, T, Barsukov, I. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras.
Nat. Cell Biol., 19, 2017
|
|
5G2Q
 
 | | The crystal structure of a S-selective transaminase from Arthrobacter sp. with alanine bound | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, TRANSAMINASE | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-04-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
4WXZ
 
 | |
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
5G39
 
 | | PsbO subunit of Photosystem II, beta barrel domain at 297K, pH 6 | | Descriptor: | CALCIUM ION, PHOTOSYSTEM II MANGANESE-STABILIZING POLYPEPTIDE | | Authors: | Bommer, M, Bondar, A.N, Zouni, A, Dobbek, H, Dau, H. | | Deposit date: | 2016-04-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic and Computational Analysis of the Barrel Part of the Psbo Protein of Photosystem II -Carboxylate-Water Clusters as Putative Proton Transfer Relays and Structural Switches
Biochemistry, 55, 2016
|
|
4X7L
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
5G3A
 
 | | PsbO subunit of Photosystem II, beta barrel domain at 100K, pH 10 | | Descriptor: | CALCIUM ION, PHOTOSYSTEM II MANGANESE-STABILIZING POLYPEPTIDE | | Authors: | Bommer, M, Bondar, A.N, Zouni, A, Dobbek, H, Dau, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.224 Å) | | Cite: | Crystallographic and Computational Analysis of the Barrel Part of the Psbo Protein of Photosystem II -Carboxylate-Water Clusters as Putative Proton Transfer Relays and Structural Switches
Biochemistry, 55, 2016
|
|
4X2H
 
 | | Sac3N peptide bound to Mex67:Mtr2 | | Descriptor: | Putative mRNA export protein, Putative uncharacterized protein, SER-SER-VAL-PHE-GLY-ALA-PRO-ALA | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Structure, 23, 2015
|
|
4X96
 
 | |
6K5I
 
 | |
5G20
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 19). | | Descriptor: | 6-(BENZYLOXY)-4-(ETHYLSULFANYL)-3-[(MORPHOLIN-4-YL), DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
7CWK
 
 | |
6UDG
 
 | |
4X4W
 
 | |
4X5W
 
 | | HLA-DR1 with CLIP102-120(M107W) | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Guenther, S, Freund, C. | | Deposit date: | 2014-12-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | MHC class II complexes sample intermediate states along the peptide exchange pathway.
Nat Commun, 7, 2016
|
|
5HAX
 
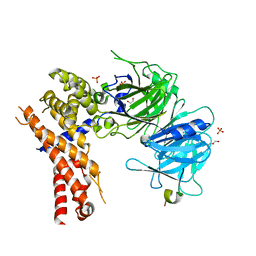 | | Crystal structure of Chaetomium thermophilum Nup170 NTD-Nup53 complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoporin NUP170, ... | | Authors: | Lin, D.H, Mobbs, G, Hoelz, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of the symmetric core of the nuclear pore.
Science, 352, 2016
|
|
5A5F
 
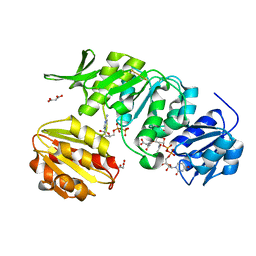 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM ESCHERICHIA COLI IN COMPLEX WITH UMA AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MALONATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE, ... | | Authors: | Sink, R, Kotnik, M, Zega, A, Barreteau, H, Gobec, S, Blanot, D, Dessen, A, Contreras-Martel, C. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of Peptidoglycan Biosynthesis Enzyme MurD: Domain Movement Revisited.
PLoS ONE, 11, 2016
|
|
5IN2
 
 | |
5I1G
 
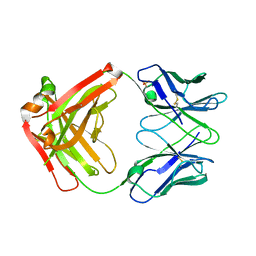 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I2K
 
 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5I0K
 
 | |
5AIE
 
 | | Not4 ring domain in complex with Ubc4 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 4, UBIQUITIN-CONJUGATING ENZYME E2 4, ZINC ION | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the Ubiquitylation Module of the Yeast Ccr4-not Complex.
Structure, 23, 2015
|
|
5A64
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine triphosphate. | | Descriptor: | 1,2-ETHANEDIOL, THIAMINE TRIPHOSPHATASE, TRIETHYLENE GLYCOL, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
