8BTU
 
 | |
8BF6
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - azotochelin complex | | Descriptor: | ABC transporter, Azotochelin, FE (III) ION, ... | | Authors: | Wilson, K.S, Duhme-Klair, A.-K, Blagova, E.V, Miller, A, Booth, R, Dodson, E.J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BS3
 
 | | Structure of USP36 in complex with Fubi-PA | | Descriptor: | 40S ribosomal protein S30, Ubiquitin carboxyl-terminal hydrolase 36, ZINC ION, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8BJ9
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - 5LICAM complex. | | Descriptor: | ABC transporter, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), ... | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BNW
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - apo form | | Descriptor: | ABC transporter, NICKEL (II) ION, SULFATE ION | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BAQ
 
 | | E. coli C7 DarT1 in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DarT ssDNA thymidine ADP-ribosyltransferase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6WPL
 
 | | Structure of Cytochrome P450tcu | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Cytochrome P-450cam, subunit of camphor 5-monooxygenase system, ... | | Authors: | Murarka, V.C, Batabyal, D, Amaya, J.A, Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Unexpected Differences between Two Closely Related Bacterial P450 Camphor Monooxygenases.
Biochemistry, 59, 2020
|
|
8BAU
 
 | | Phytophthora nicotianae var. parasitica NADAR in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, NADAR domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6WIZ
 
 | | Crystal structure of Fab 54-1G05 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
8B6I
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{S})-7-chloranyl-8-(5-methyl-2~{H}-indazol-4-yl)-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
8B7X
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - apo form. | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SULFATE ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Wilson, K.S, Duhme-Klair, A.K, Blagova, E.V, Bennett, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BAX
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - azotochelin complex. | | Descriptor: | Azotochelin, FE (III) ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Dodson, E.J, Booth, R, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6WPU
 
 | |
6WQ4
 
 | | Carbonic Anhydrase II Complexed with 2-((2-Cyanoethyl)(phenethyl)amino)-N-phenethyl-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
8BV4
 
 | | Structure of BlaC from Mycobacterium tuberculosis in complex with vaborbactam | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chikunova, A, Bruenle, S, Ubbink, M. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp179 in the class A beta-lactamase from Mycobacterium tuberculosis is a conserved yet not essential residue due to epistasis.
Febs J., 290, 2023
|
|
6WQ5
 
 | | Carbonic Anhydrase II Complexed with 2-((3-Aminopropyl)(phenethyl)amino)-N-(furan-2-ylmethyl)-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, N~2~-(3-aminopropyl)-N-[(furan-2-yl)methyl]-N~2~-(2-phenylethyl)-N-[2-(4-sulfamoylphenyl)ethyl]glycinamide, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WV4
 
 | | Human VKOR C43S with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.012 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WQ9
 
 | | Carbonic Anhydrase II Complexed with 3-((2-((Naphthalen-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WX9
 
 | | SOX2 bound to Importin-alpha 5 | | Descriptor: | Importin subunit alpha-5, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-10 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
6WXD
 
 | | SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6WYO
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) H82F F202Y double mutant complexed with Trichostatin A | | Descriptor: | Histone deacetylase 6, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.30000281 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6WXO
 
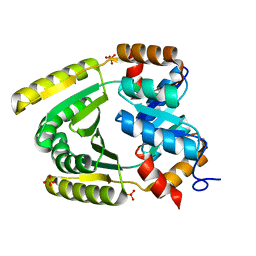 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 2-histidine 2-glutamate centre TFD-HE | | Descriptor: | GLYCEROL, SULFATE ION, TFD-HE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZ1
 
 | |
6WZQ
 
 | |
6WYF
 
 | |
