6GT6
 
 | | Crystal structure of recombinant coagulation factor beta-XIIa | | Descriptor: | CYSTEINE, Coagulation factor XII, GLYCEROL, ... | | Authors: | Pathak, M, Emsley, J. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3D6Q
 
 | | The RNase A- 5'-Deoxy-5'-N-piperidinouridine complex | | Descriptor: | 1-(5-deoxy-5-piperidin-1-yl-alpha-L-arabinofuranosyl)pyrimidine-2,4(1H,3H)-dione, CITRATE ANION, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
8AFD
 
 | | CRYSTAL STRUCTURE OF BIT-BLOCKED KRAS-G12V-S39C IN COMPLEX WITH COMPOUND 20a | | Descriptor: | (4~{S})-4-[3-(4-aminophenyl)-1,2,4-oxadiazol-5-yl]-2-azanyl-4-methyl-6,7-dihydro-5~{H}-1-benzothiophene-3-carbonitrile, 1H-benzimidazol-2-ylmethanethiol, GTPase KRas, ... | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2022-07-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
6H5Z
 
 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H79
 
 | |
3CY3
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V | | Descriptor: | (2S)-1,3-benzothiazol-2-yl{2-[(2-pyridin-3-ylethyl)amino]pyrimidin-4-yl}ethanenitrile, 1,2-ETHANEDIOL, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
To be Published
|
|
8AJ4
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A') | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], Lysozyme, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
3CYL
 
 | |
6GF9
 
 | |
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
6U5L
 
 | | Structure of human ULK4 in complex with an inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N~2~-(1H-benzimidazol-6-yl)-N~4~-(5-cyclobutyl-1H-pyrazol-3-yl)quinazoline-2,4-diamine, ... | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Resolution Structure and Inhibition of the Schizophrenia-Linked Pseudokinase ULK4.
J.Am.Chem.Soc., 142, 2020
|
|
8AJ3
 
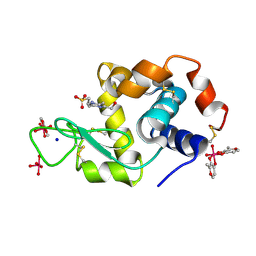 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8,8,8-tetrakis($l^{1}-oxidanyl)-2-methyl-3,7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1,5-diene, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
5DDP
 
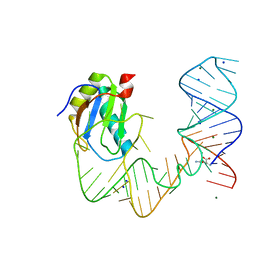 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
8AJ5
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure B) | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
5DE2
 
 | |
3CO2
 
 | |
5CWE
 
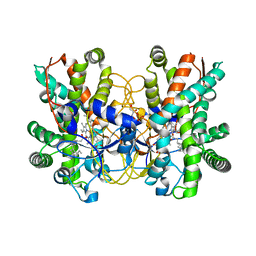 | | Structure of CYP107L2 from Streptomyces avermitilis with lauric acid | | Descriptor: | Cytochrome P450 hydroxylase, GLYCEROL, LAURIC ACID, ... | | Authors: | Pham, T.-V, Han, S.-H, Kim, J.-H, Kim, D.-H, Kang, L.-W. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of CYP107L2 from Streptomyces avermitilis with lauric acid
To Be Published
|
|
8B02
 
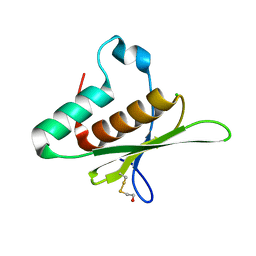 | | Crystal structure of the dsRBD domain of tRNA-dihydrouridine(20) synthase from Amphimedon queenslandica | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DRBM domain-containing protein, ... | | Authors: | Pecqueur, L, Faivre, B, Hamdane, D. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family.
Biomolecules, 12, 2022
|
|
8B3T
 
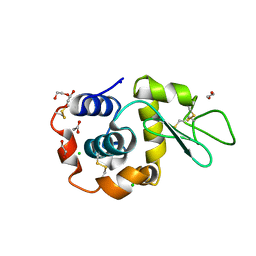 | | Hen Egg White Lysozyme 4s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
3CP4
 
 | |
3D1K
 
 | | R/T intermediate quaternary structure of an antarctic fish hemoglobin in an alpha(CO)-beta(pentacoordinate) state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, Hemoglobin subunit alpha-1, ... | | Authors: | Vitagliano, L, Vergara, A, Bonomi, G, Merlino, A, Mazzarella, L. | | Deposit date: | 2008-05-06 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.
J.Am.Chem.Soc., 130, 2008
|
|
8B3U
 
 | | Hen Egg White Lysozyme 6s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
8B3V
 
 | | Hen Egg White Lysozyme 8s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6H0K
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
