8C4X
 
 | | PAM Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nomme, J, Gavalda, S, Cioci, G, Guicherd, M, Marty, A, Duquesne, S, Ben Khaled, M. | | Deposit date: | 2023-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | An engineered enzyme embedded into PLA to make self-biodegradable plastic.
Nature, 2024
|
|
6XXQ
 
 | |
6XU2
 
 | | Human karyopherin RanBP5 (isoform-3) | | Descriptor: | Antipain, Importin-5, NICKEL (II) ION | | Authors: | Swale, C, McCarthy, A.A, Berger, I, Bieniossek, C, Delmas, B, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking.
J.Mol.Biol., 432, 2020
|
|
6XZ0
 
 | |
2XE4
 
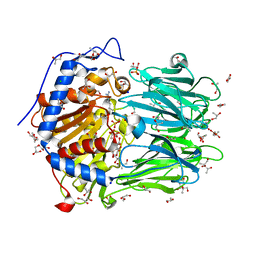 | | Structure of Oligopeptidase B from Leishmania major | | Descriptor: | ANTIPAIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | McLuskey, K, Paterson, N.G, Bland, N.D, Mottram, J.C, Isaacs, N.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Leishmania Major Oligopeptidase B Gives Insight Into the Enzymatic Properties of a Trypanosomatid Virulence Factor.
J.Biol.Chem., 285, 2010
|
|
6XTE
 
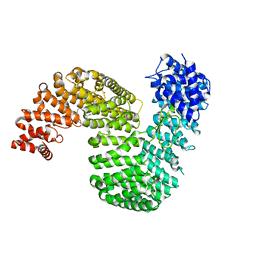 | | Human karyopherin RanBP5 (isoform-1) | | Descriptor: | Antipain, CHLORIDE ION, Importin-5, ... | | Authors: | Swale, C, McCarthy, A.A, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2020-01-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking.
J.Mol.Biol., 432, 2020
|
|
6Z28
 
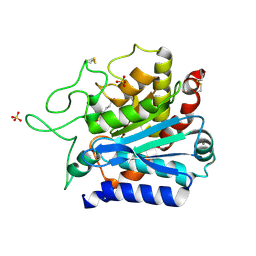 | |
5AWV
 
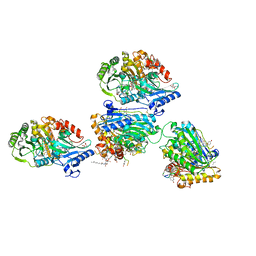 | |
8GJ4
 
 | |
8GKM
 
 | |
8GJP
 
 | |
8GIC
 
 | |
8HL8
 
 | | Crystal structrue of MtdL R257K mutant | | Descriptor: | MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
5N7Q
 
 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) IN COMPLEX WITH THE INHIBITOR PEPSTATIN A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PEPSTATIN A, ... | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-27 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
3PRT
 
 | | Mutant of the Carboxypeptidase T | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Timofeev, V.I, Akparov, V.K, Grishin, A.M, Kuranova, I.P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: |
|
|
4GM5
 
 | | Carboxypeptidase T with Sulphamoil Arginine | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-08-15 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Carboxypeptidase T with Sulphamoil Arginine
To be Published
|
|
5T3E
 
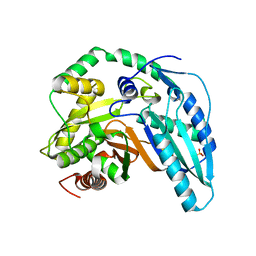 | |
1DSR
 
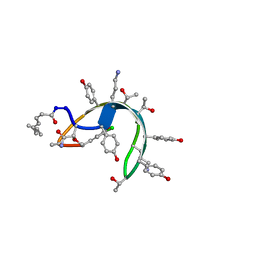 | | Peptide antibiotic, NMR, 6 structures | | Descriptor: | (2Z,4E)-7-methylocta-2,4-dienoic acid, RAMOPLANIN A2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kurz, M, Guba, W. | | Deposit date: | 1996-07-05 | | Release date: | 1997-02-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 3D Structure of Ramoplanin: A Potent Inhibitor of Bacterial Cell Wall Synthesis.
Biochemistry, 35, 1996
|
|
1JRS
 
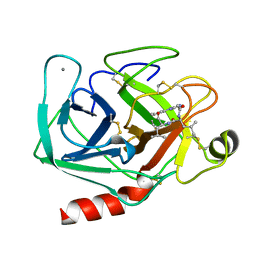 | |
1RPB
 
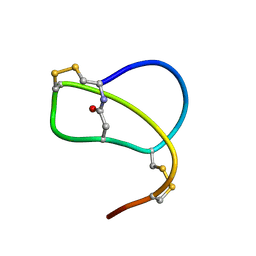 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1RPC
 
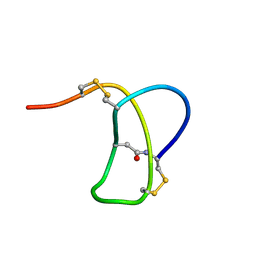 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1TL9
 
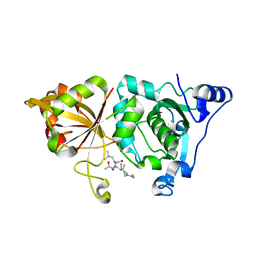 | | High resolution crystal structure of calpain I protease core in complex with leupeptin | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
1FJG
 
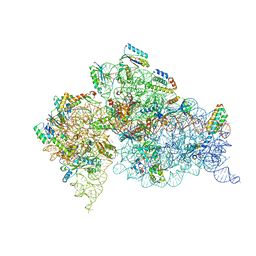 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
4BEN
 
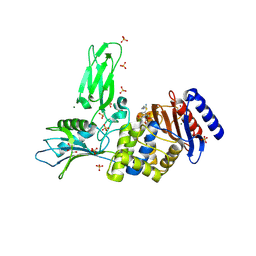 | | R39-imipenem Acyl-enzyme crystal structure | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Van Elder, D, Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-03-11 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of R39-Imipenem Acyl-Enzyme.
To be Published
|
|
4BCT
 
 | | Crystal structure of kiwi-fruit allergen Act d 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, THAUMATIN-LIKE PROTEIN | | Authors: | Pavkov-Keller, T, Bublin, M, Jankovic, M, Breiteneder, H, Keller, W. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structure of Kiwi-Fruit Allergen Act D 2
To be Published
|
|
