7PUU
 
 | | Crystal structure of carbonic anhydrase XII with methyl 4-chloro-2-cyclohexylsulfanyl-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7POM
 
 | |
7Q0D
 
 | | Human carbonic anhydrase I in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
5VQV
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7Q0E
 
 | | Human Carbonic Anhydrase II in complex with Methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUW
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7PUV
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7Q0C
 
 | | Mimic carbonic anhydrase IX in complex with Methyl 2-chloro-4-(cyclohexylsulfanyl)-5-sulfamoylbenzoate | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
3VP2
 
 | | Crystal structure of human glutaminase in complex with inhibitor 2 | | Descriptor: | 5,5'-(sulfanediyldiethane-2,1-diyl)bis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5VQQ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQY
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7PF1
 
 | | UVC treated Human apoferritin | | Descriptor: | CHLORIDE ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Renault, L, Depelteau, J.S, Briegel, A. | | Deposit date: | 2021-08-11 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | UVC inactivation of pathogenic samples suitable for cryo-EM analysis.
Commun Biol, 5, 2022
|
|
7Q6H
 
 | |
5W4Q
 
 | |
7PQV
 
 | | MEK1 IN COMPLEX WITH COMPOUND 7 | | Descriptor: | 8-(2-chloranyl-4-methoxy-phenyl)-7-fluoranyl-1-piperidin-4-yl-imidazo[4,5-c]quinoline, CALCIUM ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Moebitz, H. | | Deposit date: | 2021-09-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of MAP855, an Efficacious and Selective MEK1/2 Inhibitor with an ATP-Competitive Mode of Action.
J.Med.Chem., 65, 2022
|
|
5W4P
 
 | |
3V93
 
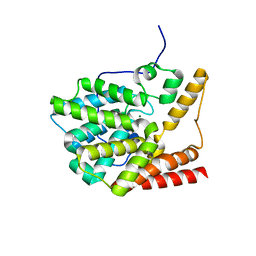 | | unliganded structure of TcrPDEC1 catalytic domain | | Descriptor: | Cyclic nucleotide specific phosphodiesterase, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Kunz, S, Chen, G, Seebeck, T, Wan, Y, Robinson, H, Martinelli, S, Ke, H. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors.
J.Biol.Chem., 287, 2012
|
|
5W4O
 
 | |
5W5V
 
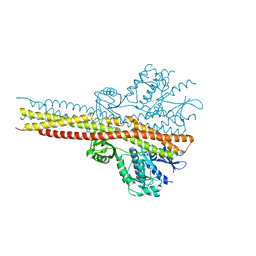 | | TBK1 co-crystal structure with amlexanox | | Descriptor: | 2-amino-7-(1-methylethyl)-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2017-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.645 Å) | | Cite: | Carboxylic Acid Derivatives of Amlexanox Display Enhanced Potency toward TBK1 and IKKepsilonand Reveal Mechanisms for Selective Inhibition.
Mol. Pharmacol., 94, 2018
|
|
3VT7
 
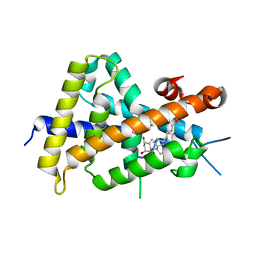 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
5WEJ
 
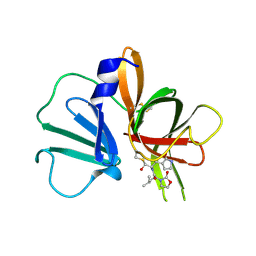 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a dipeptidyl oxazolidinone-based inhibitor | | Descriptor: | (2S)-2-{(5S)-5-[(3-chlorophenyl)methyl]-2-oxo-1,3-oxazolidin-3-yl}-4-methyl-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}pentanamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Rathnayake, A.D, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design, synthesis and evaluation of oxazolidinone-based inhibitors of norovirus 3CL protease.
Eur J Med Chem, 143, 2017
|
|
5W86
 
 | |
5WHZ
 
 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
5WBS
 
 | | Crystal structure of Frizzled-7 CRD with an inhibitor peptide Fz7-21 | | Descriptor: | Frizzled-7,inhibitor peptide Fz7-21 | | Authors: | Nile, A.H, Mukund, S, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
5WK3
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CCL17 AND M116 FAB | | Descriptor: | C-C motif chemokine 17, GLYCEROL, M116 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into chemokine CCL17 recognition by antibody M116.
Biochem Biophys Rep, 13, 2018
|
|
