8AM5
 
 | | RCII/PSI complex, class 3 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
5DF1
 
 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | Descriptor: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
6ALW
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 12-Methyl Myristic Acid (C15:0) to 1.63 Angstrom resolution | | Descriptor: | (12R)-12-methyltetradecanoic acid, (12S)-12-methyltetradecanoic acid, EDD domain protein, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
8ASP
 
 | | RCII/PSI complex, focused refinement of PSI | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
6F1W
 
 | | Crystal structure of human Casein Kinase I delta in complex with compound 31a | | Descriptor: | (9~{R},10~{R},11~{S})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
6OPA
 
 | |
9ATG
 
 | | Crystal structure of MERS 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
6FLG
 
 | |
9ATI
 
 | | Crystal structure of MERS 3CL protease in complex with a racemic bicyclo[2.2.1]heptenyl-methyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATE
 
 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptane 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9AT7
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
5DF0
 
 | | Crystal structure of AcMNPV Chitinase A in complex WITH CHITOTRIO-THIAZOLINE DITHIOAMIDE | | Descriptor: | (2R,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methylhexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
6RWD
 
 | | Crystal structure of SjGST in complex with GSH and ellagic acid at 1.53 Angstrom resolution | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Olfsen, J, Pandian, R, Sayed, Y, Dirr, H.W, Achilonu, I.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular basis of inhibition of Schistosoma japonicum glutathione transferase by ellagic acid: Insights into biophysical and structural studies.
Mol.Biochem.Parasitol., 240, 2020
|
|
1MPW
 
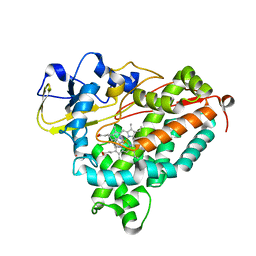 | | Molecular Recognition in (+)-a-Pinene Oxidation by Cytochrome P450cam | | Descriptor: | (+)-alpha-Pinene, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Bell, S.G, Chen, X, Sowden, R.J, Xu, F, Willams, J.N, Wong, L.-L, Rao, Z. | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam
J.Am.Chem.Soc., 125, 2003
|
|
6LKC
 
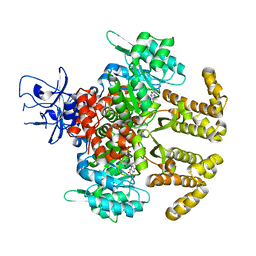 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
6TYQ
 
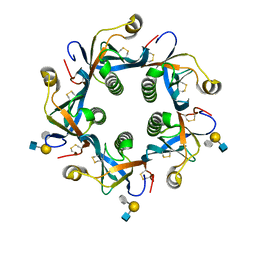 | | Salmonella Typhi PltB Homopentamer with Neu-5NAc-9OAc-alpha-2-6-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertussis-like toxin subunit B | | Authors: | Nguyen, T, Milano, S.K, Yang, Y.A, Song, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication.
Plos Pathog., 16, 2020
|
|
1A4I
 
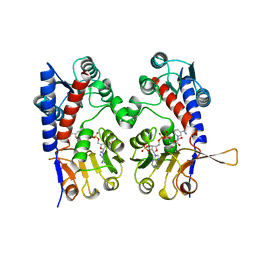 | | HUMAN TETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE / METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Allaire, M, Li, Y, Mackenzie, R.E, Cygler, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 A resolution.
Structure, 6, 1998
|
|
5DFR
 
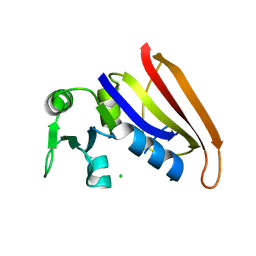 | |
6TYN
 
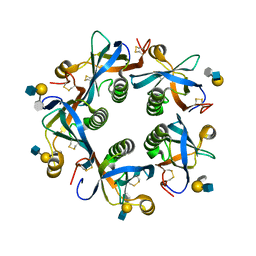 | | Salmonella Typhi PltB Homopentamer with Neu-5NAc-9OAc-alpha-2-3-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertussis like toxin subunit B | | Authors: | Nguyen, T, Milano, S.K, Yang, Y.A, Song, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication.
Plos Pathog., 16, 2020
|
|
9AU4
 
 | | Cryo-EM structure of a photosystem I variant containing an unusual plastoquinone derivative in its electron transfer chain | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Gisriel, C.J, Vasily, K, Iwig, D.F, Russell, B.P, Vinyard, D.J, Golbeck, J.H, Brudvig, G.W, Lakshmi, K.V. | | Deposit date: | 2024-02-28 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.03 Å) | | Cite: | Cryo-EM structure of a photosystem I variant containing an unusual plastoquinone derivative in its electron transfer chain.
Sci Adv, 10, 2024
|
|
6RJ5
 
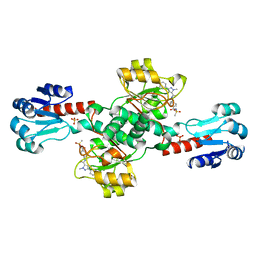 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
8V05
 
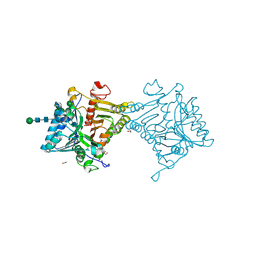 | | Crystal structure of mouse PLD3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
6AQH
 
 | |
5FS9
 
 | |
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
