3D3R
 
 | | Crystal structure of the hydrogenase assembly chaperone HypC/HupF family protein from Shewanella oneidensis MR-1 | | Descriptor: | Hydrogenase assembly chaperone hypC/hupF | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hydrogenase Assembly Chaperone HypC/HupF Family Protein from Shewanella oneidensis MR-1.
To be Published
|
|
9MF6
 
 | |
9MGJ
 
 | |
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
9MEA
 
 | |
3CRV
 
 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3D3W
 
 | |
3CSQ
 
 | |
3CTG
 
 | |
3D4U
 
 | | Bovine thrombin-activatable fibrinolysis inhibitor (TAFIa) in complex with tick-derived carboxypeptidase inhibitor. | | Descriptor: | ACETATE ION, Carboxypeptidase B2, Carboxypeptidase inhibitor, ... | | Authors: | Sanglas, L, Valnickova, Z, Arolas, J.L, Pallares, I, Guevara, T, Sola, M, Kristensen, T, Enghild, J.J, Aviles, F.X, Gomis-Ruth, F.X. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of activated thrombin-activatable fibrinolysis inhibitor, a molecular link between coagulation and fibrinolysis.
Mol.Cell, 31, 2008
|
|
6YLJ
 
 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitinhexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Endochitinase 42, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
3D54
 
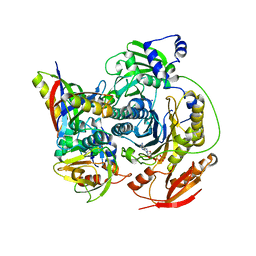 | | Structure of PurLQS from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Formylglycinamide ribonucleotide amidotransferase, Phosphoribosylformylglycinamidine synthase 1, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2008-05-15 | | Release date: | 2008-07-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation.
Biochemistry, 47, 2008
|
|
2I6Q
 
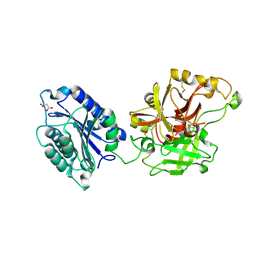 | | Complement component C2a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C2a fragment, ... | | Authors: | Milder, F.J, Raaijmakers, H.C.A, Vandeputte, D.A.A, Schouten, A, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of complement component c2a: implications for convertase formation and substrate binding.
Structure, 14, 2006
|
|
9MEC
 
 | |
3D6F
 
 | |
6YN4
 
 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitintetraose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
2I5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with DHBNH, an RNASE H inhibitor | | Descriptor: | (E)-3,4-DIHYDROXY-N'-[(2-METHOXYNAPHTHALEN-1-YL)METHYLENE]BENZOHYDRAZIDE, MAGNESIUM ION, Reverse transcriptase/ribonuclease H P51 subunit, ... | | Authors: | Himmel, D.M, Sarafianos, S.G, Knight, J.L, Levy, R.M, Arnold, E. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.
Acs Chem.Biol., 1, 2006
|
|
3D57
 
 | | TR Variant D355R | | Descriptor: | SULFATE ION, Thyroid hormone receptor beta, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Jouravel, N. | | Deposit date: | 2008-05-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for dimer formation of TRbeta variant D355R.
Proteins, 75, 2008
|
|
3CTJ
 
 | |
3D5W
 
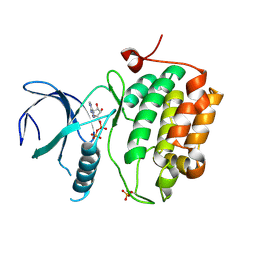 | |
3CUN
 
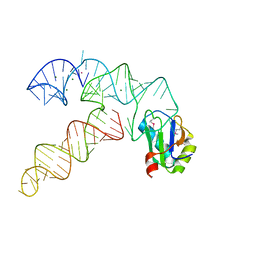 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
3CV7
 
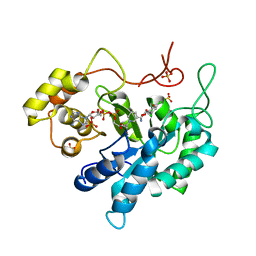 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
3D69
 
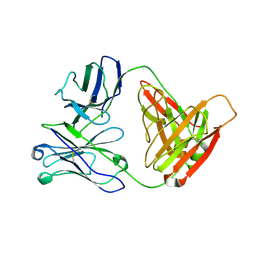 | |
5JRK
 
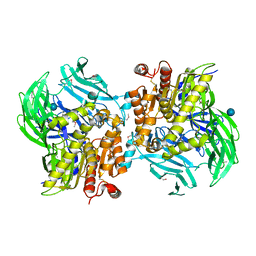 | | Crystal Structure of the Sphingopyxin I Lasso Peptide Isopeptidase SpI-IsoP (SeMet-derived) | | Descriptor: | Dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like protein, beta-D-glucopyranose | | Authors: | Fage, C.D, Hegemann, J.D, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3D7E
 
 | |
