7WNL
 
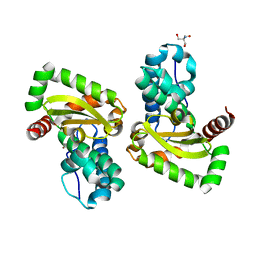 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, BROMIDE ION, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7WY2
 
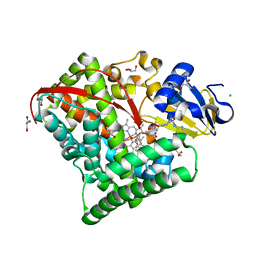 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87A Mutant Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5ZIB
 
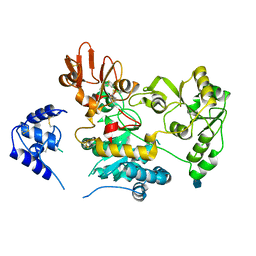 | | Crystal structure of human GnT-V luminal domain in apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
6TPO
 
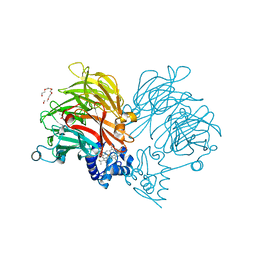 | | Conformation of cd1 nitrite reductase NirS without bound heme d1 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, HEME C, Nitrite reductase, ... | | Authors: | Kluenemann, T, Blankenfeldt, W. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structure of heme d 1 -free cd 1 nitrite reductase NirS.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8CKL
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-2)-1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
9CMV
 
 | | Crystal structure of the KRAS-p110alpha complex in the presence of molecular glue D223 | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Czyzyk, D.J, Yan, W, Simanshu, D.K. | | Deposit date: | 2024-07-15 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular glues that facilitate RAS binding to PI3K alpha promote glucose uptake without insulin.
Science, 389, 2025
|
|
3GYI
 
 | |
5F5H
 
 | | X-ray structure of Roquin ROQ domain in complex with Ox40 hexa-loop RNA motif | | Descriptor: | GLYCEROL, RNA (5'-R(P*CP*CP*AP*CP*AP*CP*CP*GP*UP*UP*CP*UP*AP*GP*GP*UP*GP*CP*UP*GP*G)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Roquin recognizes a non-canonical hexaloop structure in the 3'-UTR of Ox40.
Nat Commun, 7, 2016
|
|
5F5Z
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MA2-014 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-~{N}-[3-[[5-methyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]amino]phenyl]propane-2-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
5ZQP
 
 | | Tankyrase-2 in complex with compound 12 | | Descriptor: | 1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)-2H-spiro[1-benzofuran-3,4'-piperidin]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
7H6B
 
 | |
5F60
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SG3-014 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
1U8F
 
 | |
5SD2
 
 | |
5SD7
 
 | |
5SD1
 
 | |
5SCX
 
 | |
5FF7
 
 | |
6Q5G
 
 | |
6T77
 
 | | Crystal structure of Klebsiella pneumoniae FabG(NADPH-dependent) NADP-complex at 1.75 A resolution | | Descriptor: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
5CKX
 
 | | Non-covalent complex of DAHP synthase and chorismate mutase from Mycobacterium tuberculosis with bound transition state analog and feedback effectors tyrosine and phenylalanine | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Munack, S, Okvist, M, Krengel, U. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remote Control by Inter-Enzyme Allostery: A Novel Paradigm for Regulation of the Shikimate Pathway.
J.Mol.Biol., 428, 2016
|
|
8WGH
 
 | | Cryo-EM structure of the red-shifted Fittonia albivenis PSI-LHCI | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Huang, G.Q, Li, X.X, Sui, S.F, Qin, X.C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the red-shifted Fittonia albivenis photosystem I.
Nat Commun, 15, 2024
|
|
4OXK
 
 | | Multiple binding modes of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA within a tetramer | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8429 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
9MVO
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) in Complex with Inhibitor AVI-4692 | | Descriptor: | (3M,5P,6M)-5-(1H-1,2,3-benzotriazol-1-yl)-6-(3-chlorophenyl)-3-(isoquinolin-4-yl)-1-(prop-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione, (3M,5P,6M)-5-(1H-1,2,3-benzotriazol-1-yl)-6-(3-chlorophenyl)-3-(isoquinolin-4-yl)-1-(prop-2-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Diallo, A, Gumpena, R, Verba, K. | | Deposit date: | 2025-01-15 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-based discovery of highly bioavailable, covalent, broad-spectrum coronavirus M Pro inhibitors with potent in vivo efficacy.
Sci Adv, 11, 2025
|
|
7X8E
 
 | | Crystal structure of PfHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2022-03-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
