5LMF
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LP1
 
 | |
5LTD
 
 | | Phosphate-bound Pichia angusta Atg18 | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 18, PHOSPHATE ION | | Authors: | Scacioc, A, Kuhnel, K. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phopshate-bound Pichia angusta Atg18
To Be Published
|
|
5LY3
 
 | |
5O9S
 
 | | HsNMT1 in complex with CoA and Myristoylated-GKSNSKLK octapeptide | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5OCV
 
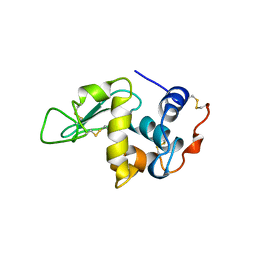 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
5NWE
 
 | | Complex of H275Y mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZ4
 
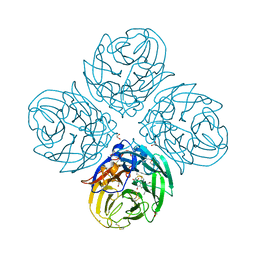 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZF
 
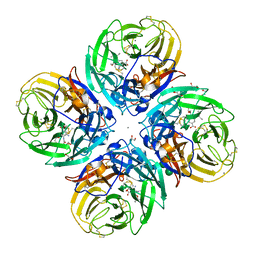 | | Complex of H275Y/I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZN
 
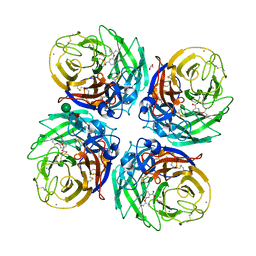 | | Complex of H275Y/S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZE
 
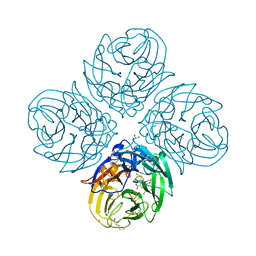 | | Complex of S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Hejdanek, J. | | Deposit date: | 2017-05-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5O9V
 
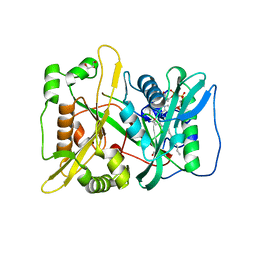 | | HsNMT1 in complex with CoA and Myristoylated-GGCFSKPK octapeptide | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5HSP
 
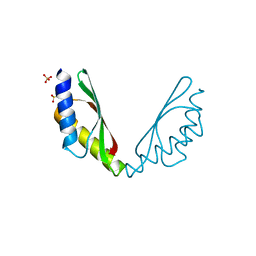 | | MamM CTD M250L | | Descriptor: | Magnetosome protein MamM, SULFATE ION | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disease-Homologous Mutation in the Cation Diffusion Facilitator Protein MamM Causes Single-Domain Structural Loss and Signifies Its Importance.
Sci Rep, 6, 2016
|
|
3D65
 
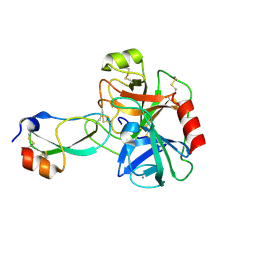 | | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Textilinin | | Authors: | Millers, E.-K.I, Masci, P.P, Lavin, M.F, de Jersey, J, Guddat, L.W. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin
To be Published
|
|
4XE4
 
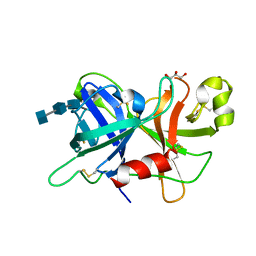 | | Coagulation Factor XII protease domain crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL | | Authors: | Pathak, M, Wilmann, P, Awford, J, Li, C, Fisher, P.M, Dreveny, I, Dekker, L.V, Emsley, J. | | Deposit date: | 2014-12-22 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coagulation factor XII protease domain crystal structure.
J.Thromb.Haemost., 13, 2015
|
|
7RX4
 
 | | Cryo-EM reconstruction of AS2 nanotube (Form II like) | | Descriptor: | AS2 peptide | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
7RX5
 
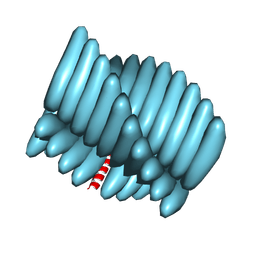 | | Cryo-EM reconstruction of Form1-N2 nanotube (Form I like) | | Descriptor: | F1-N2 nanotube | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
6A98
 
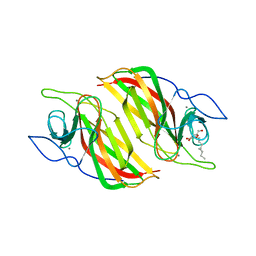 | | Crystal structure of a cyclase from Fischerella sp. TAU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, aromatic prenyltransferase, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8BQU
 
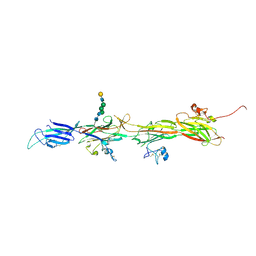 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
6A6B
 
 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
6A8X
 
 | | Crystal structure of a apo form cyclase from Fischerella sp. | | Descriptor: | CALCIUM ION, aromatic prenyltransferase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7S64
 
 | |
7S63
 
 | |
7S62
 
 | |
8BH3
 
 | | DNA-PK Ku80 mediated dimer bound to PAXX | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Hardwick, S.W, Chaplin, A.K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
