6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
6HRD
 
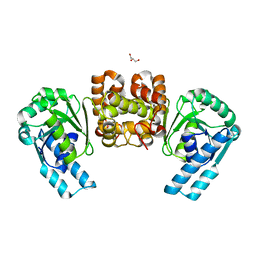 | |
6F9G
 
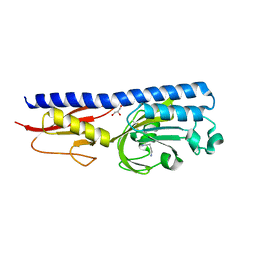 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
5E9O
 
 | | Spirochaeta thermophila X module - CBM64 - mutant G504A | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Cellulase, glycosyl hydrolase family 5, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cellulose binding by the type A carbohydrate-binding module 64 of Spirochaeta thermophila.
Proteins, 84, 2016
|
|
7RH6
 
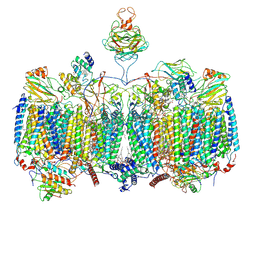 | | Mycobacterial CIII2CIV2 supercomplex, inhibitor free, -Lpqe cyt cc open | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Di Trani, J.M, Yanofsky, D.J, Rubinstein, J.L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial CIII 2 CIV 2 respiratory supercomplex bound to the tuberculosis drug candidate telacebec (Q203).
Elife, 10, 2021
|
|
7RH5
 
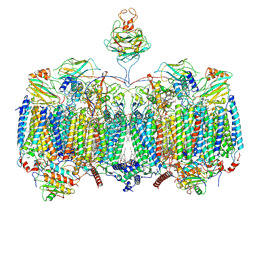 | | Mycobacterial CIII2CIV2 supercomplex, Inhibitor free | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Di Trani, J.M, Yanofsky, D.J, Rubinstein, J.L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-04 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of mycobacterial CIII 2 CIV 2 respiratory supercomplex bound to the tuberculosis drug candidate telacebec (Q203).
Elife, 10, 2021
|
|
9H25
 
 | |
2N9N
 
 | |
9H26
 
 | | Structure of rsCherry exposed to oxygen for 69 days | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2024-10-11 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure analysis of oxygen-induced degradation occurring in rsCherry.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
5N16
 
 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 1) | | Descriptor: | 5-cyclopropyl-2-(5-pyrazin-2-yl-1,2,4-oxadiazol-3-yl)benzo[b][1,4]benzothiazepin-6-one, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
1M7X
 
 | | The X-ray Crystallographic Structure of Branching Enzyme | | Descriptor: | 1,4-alpha-glucan Branching Enzyme | | Authors: | Abad, M.C, Binderup, K, Rios-Steiner, J, Arni, R.K, Preiss, J, Geiger, J.H. | | Deposit date: | 2002-07-23 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystallographic structure of Escherichia coli branching enzyme
J.Biol.Chem., 277, 2002
|
|
5CVR
 
 | |
6FBE
 
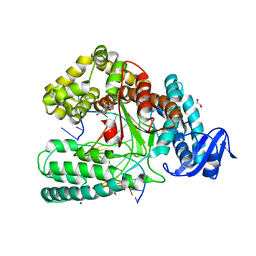 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the third primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*GP*GP*TP*GP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GZY
 
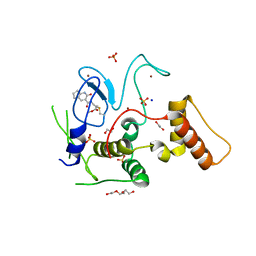 | | HOIP-fragment5 complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF31, SODIUM ION, ... | | Authors: | Johansson, H, Tsai, Y.C.I, Fantom, K, Chung, C.W, Martino, L, House, D, Rittinger, K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP.
J. Am. Chem. Soc., 141, 2019
|
|
5FXE
 
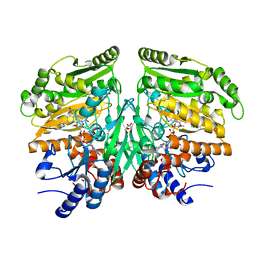 | | Crystal structure of eugenol oxidase in complex with coniferyl alcohol | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, EUGENOL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nguyen, Q.-T, de Gonzalo, G, Binda, C, Martinez, A.R, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Eugenol Oxidase from Rhodococcus Jostii Rha1: A Versatile Oxidative Biocatalyst.
Chembiochem, 17, 2016
|
|
8XGY
 
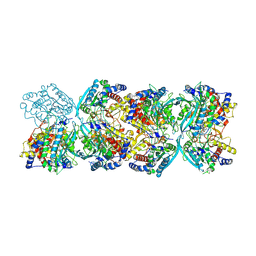 | |
8OVD
 
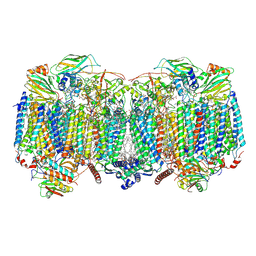 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
9H02
 
 | | Crystal structure of human CREBBP histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Mechaly, A.E, Cui, G, Green, M.R, Rodrigues-Lima, F. | | Deposit date: | 2024-10-07 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of human CREBBP histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8RYU
 
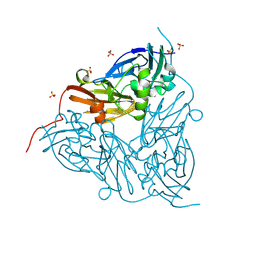 | | High pH (8.0) nitrite-bound MSOX movie series dataset 10 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [6.9 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RYV
 
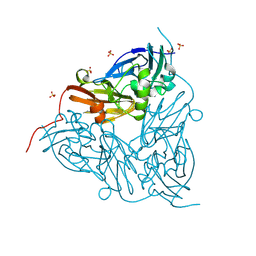 | | MSOX movie series dataset 10 (11.5 MGy) for nitrite bound BrJNiR (Cu containing nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 at pH 8. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RYR
 
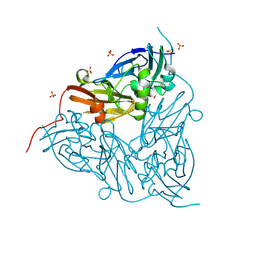 | | High pH (8.0) nitrite-bound MSOX movie series dataset 4 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [2.76 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8RYJ
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 3 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [2.07 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7UMX
 
 | | Crystal structure of Acinetobacter baumannii FabI in complex with NAD and (R,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide | | Descriptor: | (2E)-3-[(7R)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hajian, B. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
5N1E
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule N-(1,3-benzodioxol-5-yl)-2-piperidin-1-ylacetamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | A crystallographic fragment study with cAMP-dependent protein kinase A
To Be Published
|
|
6HPI
 
 | |
