7DBL
 
 | | Acyl-CoA hydrolase MpaH' mutant S139A in complex with MPA | | Descriptor: | MYCOPHENOLIC ACID, acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
1FJ4
 
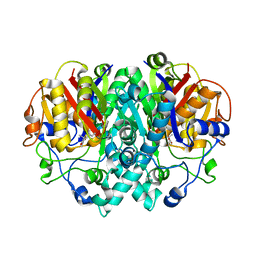 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1VFU
 
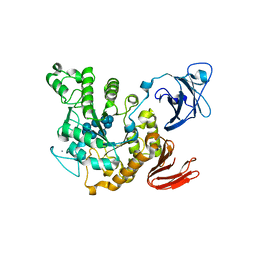 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFM
 
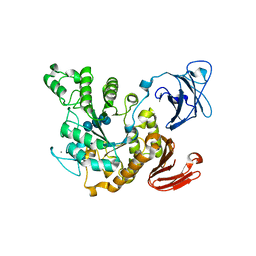 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/alpha-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
5DOY
 
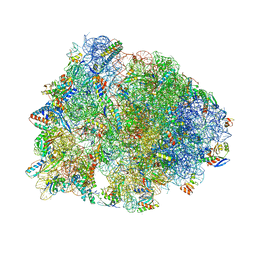 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic Hygromycin A, mRNA and three tRNAs in the A, P and E sites at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
4JH3
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and Fosfomycin | | Descriptor: | FORMIC ACID, FOSFOMYCIN, GLYCEROL, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
7ZE9
 
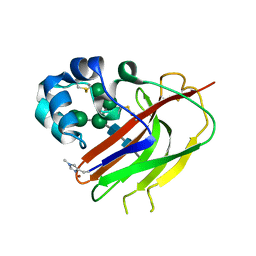 | | Structure of an AA16 LPMO-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Huang, Z, Banerjee, S, Muderspach, S.J, Sun, P, van Berkel, W.J.H, Kabel, M.A, Lo Leggio, L. | | Deposit date: | 2022-03-30 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | AA16 Oxidoreductases Boost Cellulose-Active AA9 Lytic Polysaccharide Monooxygenases from Myceliophthora thermophila.
Acs Catalysis, 13, 2023
|
|
4JH5
 
 | | Crystal Structure of FosB from Bacillus cereus with Cobalt and Fosfomycin | | Descriptor: | COBALT (II) ION, FOSFOMYCIN, MAGNESIUM ION, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
4JH8
 
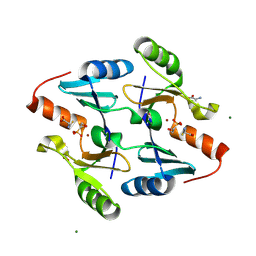 | | Crystal Structure of FosB from Bacillus cereus with Zinc and L-Cysteine-Fosfomycin Ternary Complex | | Descriptor: | CYSTEINE, FOSFOMYCIN, GLYCEROL, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
2VCD
 
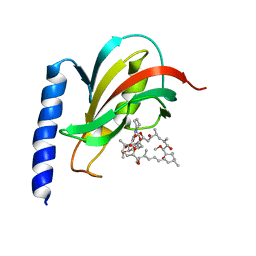 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
1YFC
 
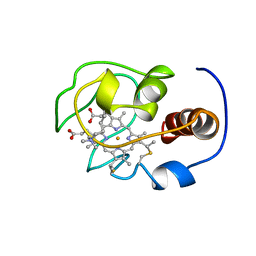 | | Solution nmr structure of a yeast iso-1-ferrocytochrome C | | Descriptor: | HEME C, YEAST ISO-1-FERROCYTOCHROME C | | Authors: | Baistrocchi, P, Banci, L, Bertini, I, Turano, P, Bren, K.L, Gray, H.B. | | Deposit date: | 1996-08-08 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c.
Biochemistry, 35, 1996
|
|
6SE7
 
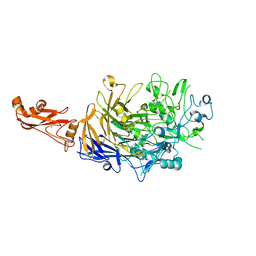 | |
1JR1
 
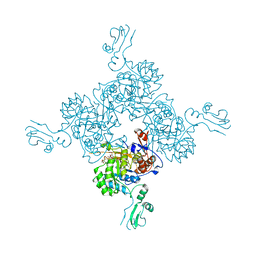 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
1KAN
 
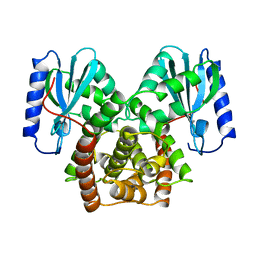 | |
2D2O
 
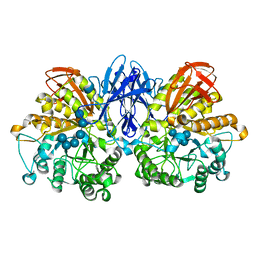 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
2GFI
 
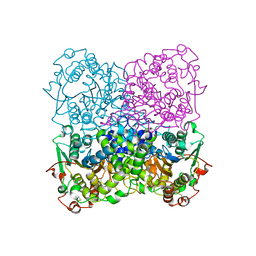 | | Crystal structure of the phytase from D. castellii at 2.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Hoh, F. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Debaryomyces castellii CBS 2923 phytase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5FB7
 
 | |
5ZI9
 
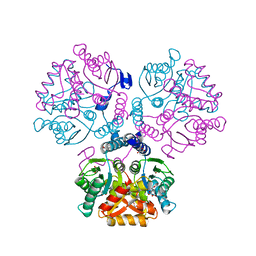 | | Crystal structure of type-II LOG from Streptomyces coelicolor A3 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, ... | | Authors: | Seo, H, Kim, K.-J. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical characterization of the type-II LOG protein from Streptomyces coelicolor A3.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
3A6O
 
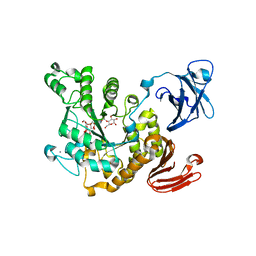 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/acarbose complex | | Descriptor: | ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
4JAX
 
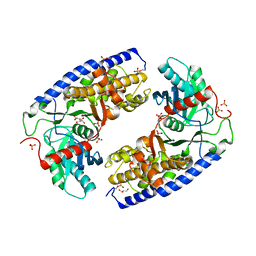 | | Crystal structure of dimeric KlHxk1 in crystal form X | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Strater, N, Kettner, K, Otto, A, Lilie, H, Golbik, R.P, Kriegel, T.M. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | In vivo phosphorylation and in vitro autophosphorylation-inactivation of Kluyveromyces lactis hexokinase KlHxk1.
Biochem.Biophys.Res.Commun., 435, 2013
|
|
4KR8
 
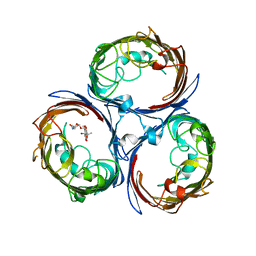 | |
5YSW
 
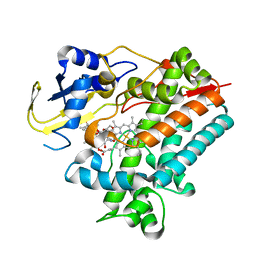 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
7N7G
 
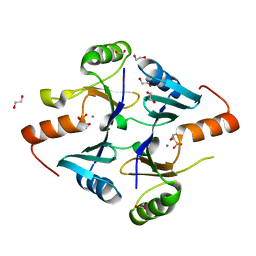 | | Crystal Structure of FosB from Enterococcus faecium with Fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, MANGANESE (II) ION, ... | | Authors: | Shay, M.R, Simmons, Z, Thompson, M.K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of fosfomycin resistance conferred by FosB from Enterococcus faecium.
Protein Sci., 31, 2022
|
|
3U14
 
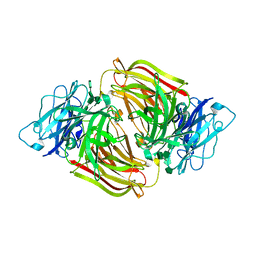 | | Structure of D50A-fructofuranosidase from Schwanniomyces occidentalis complexed with inulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
3U75
 
 | | Structure of E230A-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosylnystose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
