1P1R
 
 | | Horse liver alcohol dehydrogenase complexed with NADH and R-N-1-methylhexylformamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R)-N-(1-METHYL-HEXYL)-FORMAMIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Venkataramaiah, T.H, Plapp, B.V. | | Deposit date: | 2003-04-13 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Formamides mimic aldehydes and inhibit liver alcohol dehydrogenases and ethanol metabolism
J.Biol.Chem., 278, 2003
|
|
1O2D
 
 | |
4EML
 
 | |
4AMX
 
 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-glucosyl- fluoride | | Descriptor: | 5-fluoro-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1O9J
 
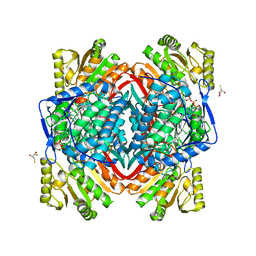 | | The X-ray crystal structure of eta-crystallin | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ALDEHYDE DEHYDROGENASE, ... | | Authors: | Purkiss, A.G, Van Montfort, R, Wistow, G, Slingsby, C. | | Deposit date: | 2002-12-15 | | Release date: | 2003-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Eta-Crystallin: Adaptation of a Class 1 Aldehyde Dehydrogenase for a New Role in the Eye Lens
Biochemistry, 42, 2003
|
|
2J9J
 
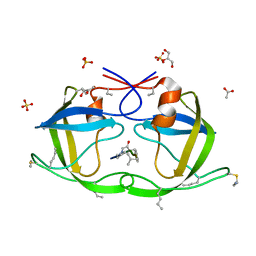 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
1HQS
 
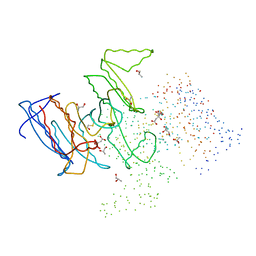 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
7JHM
 
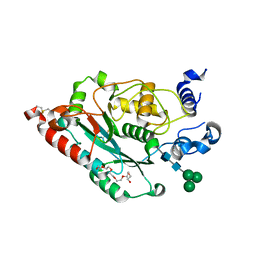 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
3QVT
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus wild-type with the intermediate 5-keto 1-phospho glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
2JE4
 
 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
1ARP
 
 | |
5CJX
 
 | |
7JHN
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP and trisaccharide GlcNAc-beta1-3Gal-beta1-4GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
4HFR
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with an orally bioavailable acidic inhibitor AZD4017. | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {(3S)-1-[5-(cyclohexylcarbamoyl)-6-(propylsulfanyl)pyridin-2-yl]piperidin-3-yl}acetic acid | | Authors: | Ogg, D.J, Gerhardt, S, Hargreaves, D. | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Bioavailable Acidic 11 -Hydroxysteroid Dehydrogenase Type 1 (11 -HSD1) Inhibitor: Discovery of 2-[(3S)-1-[5-(Cyclohexylcarbamoyl)-6-propylsulfanylpyridin-2-yl]-3-piperidyl]acetic Acid (AZD4017)
J.Med.Chem., 55, 2012
|
|
4BG1
 
 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 1-(3-Carboxypropyl)-1-methylpyrrolidin-1-ium chloride | | Descriptor: | 1-(3-carboxypropyl)-1-methylpyrrolidin-1-ium, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
2GH5
 
 | | Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5 | | Descriptor: | 6-(3-METHYL-1,4-DIOXO-1,4-DIHYDRONAPHTHALEN-2-YL)HEXANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Winzer, A, Bauer, H, Schirmer, H, Davioud-Charvet, E. | | Deposit date: | 2006-03-25 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
J.Am.Chem.Soc., 128, 2006
|
|
2OTY
 
 | | 1,2-dichlorobenzene in complex with T4 Lysozyme L99A | | Descriptor: | 1,2-DICHLOROBENZENE, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Graves, A.P, Shoichet, B.K. | | Deposit date: | 2007-02-09 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Predicting absolute ligand binding free energies to a simple model site.
J.Mol.Biol., 371, 2007
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
2OU0
 
 | |
1EQU
 
 | | TYPE 1 17-BETA HYDROXYSTEROID DEHYDROGENASE EQUILIN COMPLEXED WITH NADP+ | | Descriptor: | EQUILIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (ESTRADIOL 17 BETA-DEHYDROGENASE 1) | | Authors: | Sawicki, M.W, Erman, M, Puranen, T, Vihko, P, Ghosh, D. | | Deposit date: | 1998-12-02 | | Release date: | 1999-12-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the ternary complex of human 17beta-hydroxysteroid dehydrogenase type 1 with 3-hydroxyestra-1,3,5,7-tetraen-17-one (equilin) and NADP+.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RF9
 
 | | Crystal structure of cytochrome P450-cam with a fluorescent probe D-4-AD (Adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-BUTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational States of Cytochrome P450cam Revealed by Trapping of synthetic Molecular Wires
J.Mol.Biol., 344, 2004
|
|
3QQP
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1B8G
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
5TIP
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TIQ
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
