4DT4
 
 | |
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRP
 
 | | Evaluation of Synthetic FK506 Analogs as Ligands for the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-3-(3,4-dimethoxyphenyl)-1-((S)-1-(2-((1R,2S)-2-ethyl-1-hydroxy-cyclohexyl)-2-oxoacetyl)piperidine-2-carbonyloxy)propyl)phenoxy)acetic acid from cocrystallization | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRN
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRM
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRK
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRJ
 
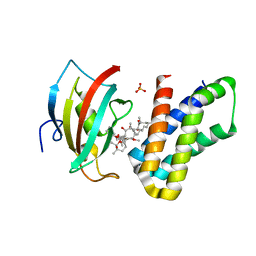 | | o-crystal structure of the PPIase domain of FKBP52, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRI
 
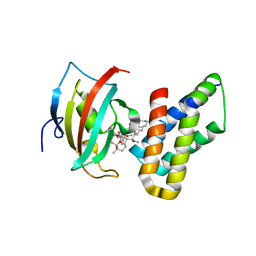 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRH
 
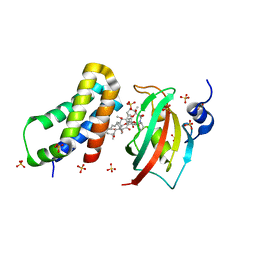 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR at low pH | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DIP
 
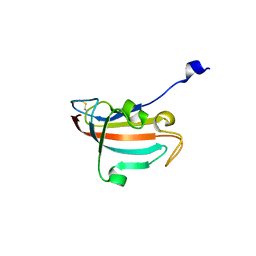 | | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14 | | Descriptor: | PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase FKBP14, SODIUM ION | | Authors: | Krojer, T, Kiyani, W, Goubin, S, Muniz, J.R.C, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Oppermann, U, Zschocke, J, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14
To be Published
|
|
4DH0
 
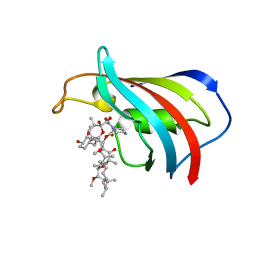 | |
4C02
 
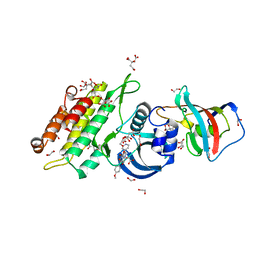 | | Crystal structure of human ACVR1 (ALK2) in complex with FKBP12.6 and dorsomorphin | | Descriptor: | 1,2-ETHANEDIOL, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Williams, E, Riesebos, E, Vollmar, M, Krojer, T, Bradley, A, Shrestha, L, Kupinska, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of Human Acvr1 (Alk2) in Complex with Fkbp12.6 And Dorsomorphin
Ph D Thesis
|
|
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
3VAW
 
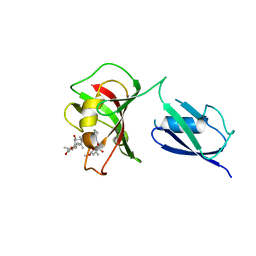 | |
3UQI
 
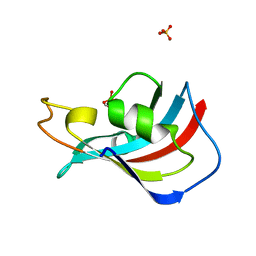 | |
3UQB
 
 | |
3UQA
 
 | |
3UF8
 
 | |
3PRD
 
 | |
3PRB
 
 | |
3PRA
 
 | |
3PR9
 
 | |
3PA7
 
 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
3O5R
 
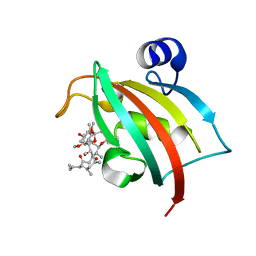 | | Complex of Fk506 with the Fk1 domain mutant A19T of FKBP51 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
