8GLM
 
 | |
6WOT
 
 | | Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
6X36
 
 | | Pig R615C RyR1 in complex with CaM, EGTA (class 3, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6WOU
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 mutant R176Q in complex with FKBP12.6 in nanodisc | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
8G01
 
 | | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase SlyD, GPE, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Riera, N. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
8G02
 
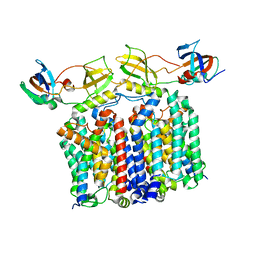 | | YES Complex - E. coli MraY, Protein E PhiX174, E. coli SlyD | | Descriptor: | Lysis protein E, Peptidyl-prolyl cis-trans isomerase, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Li, Y.E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
6X32
 
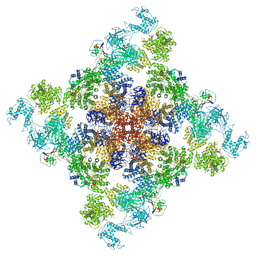 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 1 and 2, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
1FAP
 
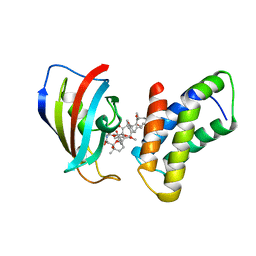 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
1Q6H
 
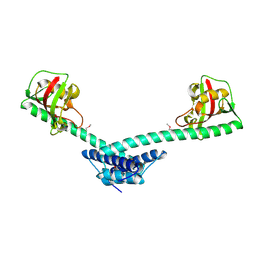 | | Crystal structure of a truncated form of FkpA from Escherichia coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6U
 
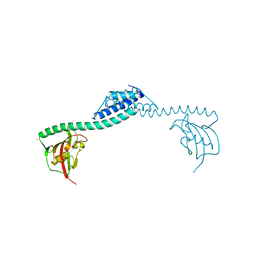 | | Crystal structure of FkpA from Escherichia coli | | Descriptor: | CESIUM ION, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-14 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6I
 
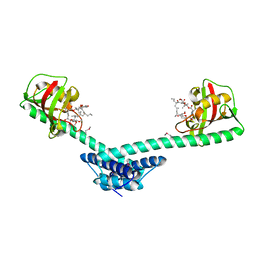 | | Crystal structure of a truncated form of FkpA from Escherichia coli, in complex with immunosuppressant FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1QZ2
 
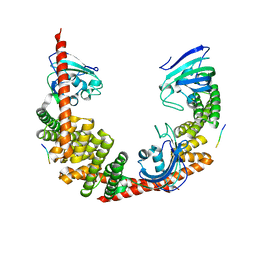 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6H3J
 
 | |
6H3I
 
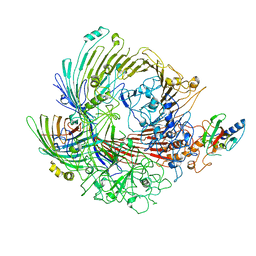 | | Structural snapshots of the Type 9 protein translocon | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, PorV, Protein involved in gliding motility SprA | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2018-07-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Type 9 secretion system structures reveal a new protein transport mechanism.
Nature, 564, 2018
|
|
6JIU
 
 | | Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JI8
 
 | | Structure of RyR2 (F/apoCaM dataset) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JII
 
 | | Structure of RyR2 (F/A/C/L-Ca2+/apo-CaM-M dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JIY
 
 | | Structure of RyR2 (F/A/C/H-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-24 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
1T11
 
 | | Trigger Factor | | Descriptor: | Trigger factor | | Authors: | Ludlam, A.V, Moore, B.A, Xu, Z. | | Deposit date: | 2004-04-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8JGA
 
 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
3PRD
 
 | |
3PRA
 
 | |
2MPH
 
 | |
3PR9
 
 | |
3PRB
 
 | |
