4ORC
 
 | | Crystal structure of mammalian calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
4Q7F
 
 | | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine. | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 5' nucleotidase family protein, MAGNESIUM ION, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine.
TO BE PUBLISHED
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ORB
 
 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
3H67
 
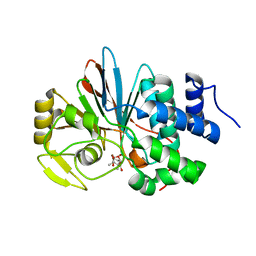 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c)with two Zn2+ atoms complexed with cantharidic acid | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, Serine/threonine-protein phosphatase 5, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
3H62
 
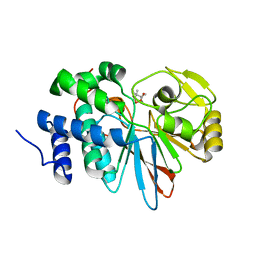 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Mn2+ atoms complexed with cantharidic acid | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
3H69
 
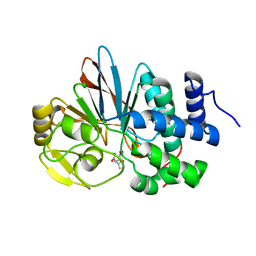 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Zn2+ atoms complexed with endothall | | Descriptor: | (1R,2S,3R,4S)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, Serine/threonine-protein phosphatase 5, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
3H68
 
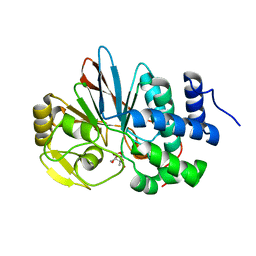 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c)with two Zn2+ atoms originally soaked with cantharidin (which is present in the structure in the hydrolyzed form) | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, Serine/threonine-protein phosphatase 5, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
5FCB
 
 | | Murine SMPDL3A in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FI9
 
 | | Closed form of murine Acid Sphingomyelinase in complex with bisphosphonate inhibitor AbPA | | Descriptor: | (1-azanyl-1-phosphono-decyl)phosphonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
3FGA
 
 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
5HQN
 
 | | Catalytic domain of murine Acid Sphingomyelinase (ASMase, ASM, SMPD1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
5HPE
 
 | | Phosphatase domain of PP5 bound to a phosphomimetic Cdc37 substrate peptide | | Descriptor: | COBALT HEXAMMINE(III), MANGANESE (II) ION, Serine/threonine-protein phosphatase 5,Hsp90 co-chaperone Cdc37 | | Authors: | Oberoi, J, Mariotti, L, Vaughan, C. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and functional basis of protein phosphatase 5 substrate specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5FCA
 
 | | Murine SMPDL3A in presence of excess zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FC7
 
 | | Murine SMPDL3A in complex with sulfate (tetragonal) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FC1
 
 | | Murine SMPDL3A in complex with sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5FIB
 
 | | Open form of murine Acid Sphingomyelinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
5INB
 
 | | RepoMan-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
5H7W
 
 | |
5FC6
 
 | | Murine SMPDL3A in complex with ADP analog AMPCP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
3ICF
 
 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
3H64
 
 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Mn2+ atoms complexed with endothall | | Descriptor: | (1R,2S,3R,4S)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
5FIC
 
 | | Open form of murine Acid Sphingomyelinase in presence of lipid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mammalian acid sphingomyelinase.
Nat Commun, 7, 2016
|
|
