2PBW
 
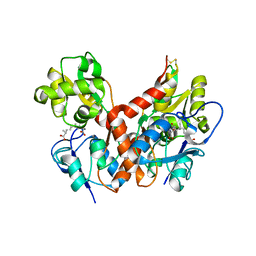 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
3HO6
 
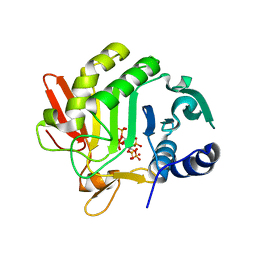 | |
3I06
 
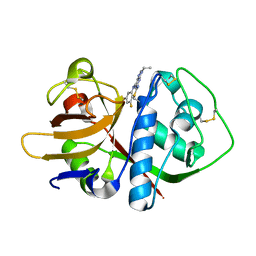 | | Crystal structure of cruzain covalently bound to a purine nitrile | | Descriptor: | 6-[(3,5-difluorophenyl)amino]-9-ethyl-9H-purine-2-carbonitrile, Cruzipain | | Authors: | Ferreira, R.S, Shoichet, B.K, McKerrow, J.H. | | Deposit date: | 2009-06-24 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and optimization of inhibitors of trypanosomal cysteine proteases: cruzain, rhodesain, and TbCatB.
J.Med.Chem., 53, 2010
|
|
7SNB
 
 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Fatty Acid Kinase A, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution
To Be Published
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7RES
 
 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TE9
 
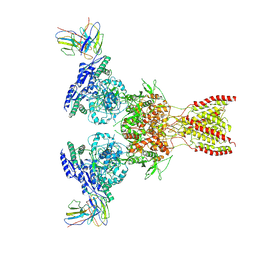 | |
2H7S
 
 | | L244A mutant of Cytochrome P450cam | | Descriptor: | Cytochrome P450-cam, HEME C | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
7BGF
 
 | |
7T0V
 
 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
2H66
 
 | | The Crystal Structure of Plasmodium Vivax 2-Cys peroxiredoxin | | Descriptor: | PV-PF14_0368 | | Authors: | Wernimont, A.K, Dong, A, Zhao, Y, Lew, J, Melone, M, Kozieradzki, I, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
7RGL
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5MHI
 
 | |
3HQP
 
 | | Crystal structure of Leishmania mexicana pyruvate kinase (LmPYK) in complex with ATP, Oxalate and fructose 2,6 bisphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Morgan, H.P, Walkinshaw, M.D. | | Deposit date: | 2009-06-08 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric mechanism of pryuvate kinase from Leishmania mexicana: a rock and lock model
J.Biol.Chem., 285, 2010
|
|
8RDU
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LE, MAGNESIUM ION, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
7LOX
 
 | | The structure of Agmatinase from E. Coli at 3.2 A displaying guanidine in the active site | | Descriptor: | Agmatinase, GUANIDINE, MANGANESE (II) ION | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
2H92
 
 | |
7T2Z
 
 | |
7LYG
 
 | |
2H9N
 
 | | WDR5 in complex with monomethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
7RFI
 
 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7LDE
 
 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
5MJ2
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form after UV-RIP (S-SAD data) | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
7LDD
 
 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
2OZE
 
 | | The Crystal structure of Delta protein of pSM19035 from Streptoccocus pyogenes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Orf delta', ... | | Authors: | Cicek, A, Weihofen, W, Saenger, W. | | Deposit date: | 2007-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation.
Nucleic Acids Res., 36, 2008
|
|
