6NHK
 
 | | Mortalin nucleotide binding domain in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Page, R.C, Moseng, M.A, Nix, J.C. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.777 Å) | | Cite: | Biophysical Consequences of EVEN-PLUS Syndrome Mutations for the Function of Mortalin.
J.Phys.Chem.B, 123, 2019
|
|
6ND9
 
 | |
4NE1
 
 | | Human MHF1 MHF2 DNA complexes | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NFF
 
 | | Human kallikrein-related peptidase 2 in complex with PPACK | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Kallikrein-2 | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
6NDR
 
 | |
6NKP
 
 | |
4NIP
 
 | |
6NFV
 
 | | Structure of the KcsA-G77C mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DO4
 
 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
6NTI
 
 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NBO
 
 | |
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
4NLV
 
 | | Poliovirus Polymerase - G289A/C290F Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
6NCS
 
 | |
4NNT
 
 | |
4NCC
 
 | | Neutralizing antibody to murine norovirus | | Descriptor: | Fab fragment heavy, Fab fragment light | | Authors: | Smith, T, Li, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization.
J.Virol., 88, 2014
|
|
6NDE
 
 | |
6NE2
 
 | | Designed repeat protein in complex with Fz7 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6DPX
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (3-{[(3-chloro-2-hydroxyphenyl)sulfonyl]amino}phenyl)acetic acid, Beta-lactamase | | Authors: | Singh, I. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ultra-large library docking for discovering new chemotypes.
Nature, 566, 2019
|
|
6NE9
 
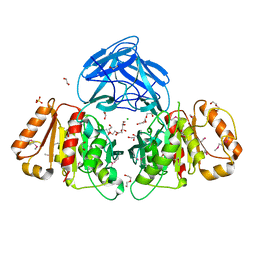 | | Bacteroides intestinalis acetyl xylan esterase (BACINT_01039) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
4NDS
 
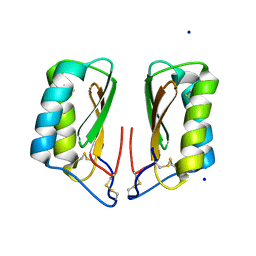 | |
4NE3
 
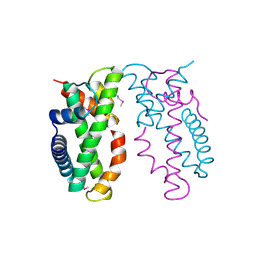 | | Human MHF1-MHF2 complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8007 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
4URK
 
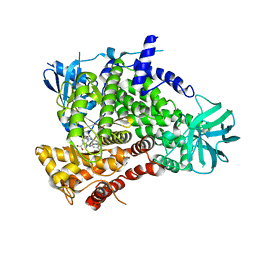 | | PI3Kg in complex with AZD6482 | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6NK4
 
 | |
