8P5F
 
 | | Human wild-type GAPDH,orthorhombic form | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Samygina, V.R, Muronetz, V.I, Schmalhausen, E.V. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-nitrosylation and S-glutathionylation of GAPDH: Similarities, differences, and relationships.
Biochim Biophys Acta Gen Subj, 1867, 2023
|
|
5ZJK
 
 | | Structure of myroilysin | | Descriptor: | Myroilysin, PHOSPHATE ION, ZINC ION | | Authors: | Li, W.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mature myroilysin and implication for its activation mechanism.
Int.J.Biol.Macromol., 2019
|
|
6BBT
 
 | | Structure of the major pilin protein (T-13) from Streptococcus pyogenes serotype GAS131465 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Young, P.G, Baker, E.N, Moreland, N.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
1JD2
 
 | | Crystal Structure of the yeast 20S Proteasome:TMC-95A complex: A non-covalent Proteasome Inhibitor | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Koguchi, Y, Huber, R, Kohno, J. | | Deposit date: | 2001-06-12 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the 20 S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor.
J.Mol.Biol., 311, 2001
|
|
4HSS
 
 | |
6WQL
 
 | |
6WQJ
 
 | |
4NSV
 
 | | Lysobacter enzymogenes lysc endoproteinase K30R mutant covalently inhibited by TLCK | | Descriptor: | CHLORIDE ION, Lysyl endopeptidase, N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-4-methylbenzenesulfonamide (Bound Form), ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7QTZ
 
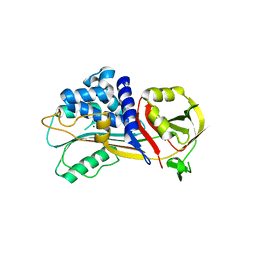 | | Crystal structure of Iripin-1 serpin from tick Ixodes ricinus | | Descriptor: | MAGNESIUM ION, Putative salivary serpin | | Authors: | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.
Front Immunol, 14, 2023
|
|
8VC3
 
 | | Voltage gated potassium ion channel Kv1.2 in complex with DTx | | Descriptor: | Kunitz-type serine protease inhibitor homolog alpha-dendrotoxin, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2 | | Authors: | Wu, Y, Sigworth, F.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Kv1.2 potassium channels, conducting and non-conducting.
Biorxiv, 2024
|
|
8WVB
 
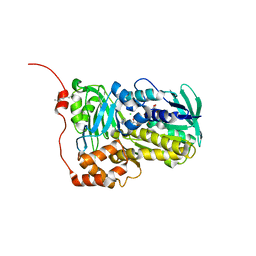 | | Crystal structure of Lsd18 mutant S195M | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WVF
 
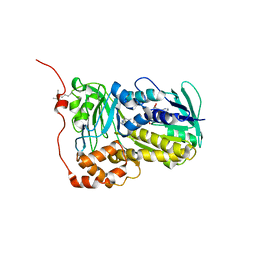 | | Crystal structure of Lsd18 mutant T189M and S195M | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.757 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
4NSY
 
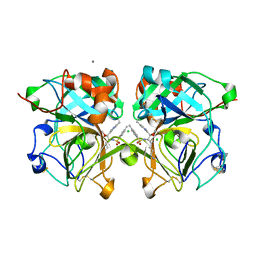 | | Wild-type lysobacter enzymogenes lysc endoproteinase covalently inhibited by TLCK | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lysyl endopeptidase, ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6WDW
 
 | |
6WDY
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Indole Phenylhydroxamate Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-hydroxy-4-[(1H-indol-1-yl)methyl]benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
4MOA
 
 | | Crystal structure of CRY4BA-R203Q TOXIN | | Descriptor: | Pesticidal crystal protein cry4Ba | | Authors: | Thamwiriyasati, N, Angsuthanasombat, C, Chen, C.-J. | | Deposit date: | 2013-09-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potential Prepore Trimer Formation by the Bacillus thuringiensis Mosquito-specific Toxin: MOLECULAR INSIGHTS INTO A CRITICAL PREREQUISITE OF MEMBRANE-BOUND MONOMERS
J.Biol.Chem., 290, 2015
|
|
1RYP
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
4K22
 
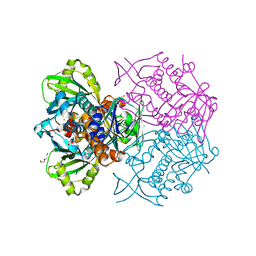 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
9CMI
 
 | |
5JVK
 
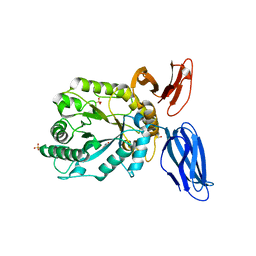 | | Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Ali-Ahmad, A, Garron, M.L, Zamboni, V, Lenfant, N, Bourne, Y, Vincent, F. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2.
J. Struct. Biol., 197, 2017
|
|
4QQG
 
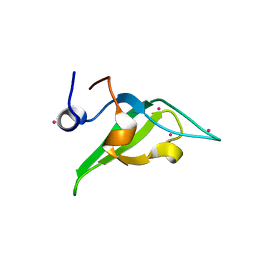 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
5KCH
 
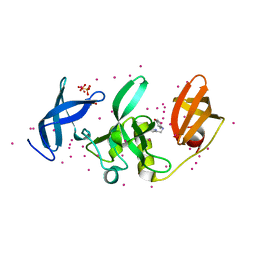 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy into weak electron density | | Descriptor: | 4-methoxy-N-[(pyridin-2-yl)methyl]aniline, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KH6
 
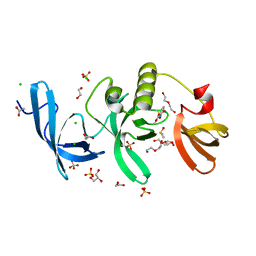 | | SETDB1 in complex with a fragment candidate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Walker, J.R, Harding, R.J, Mader, P, Dobrovetsky, E, Dong, A, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Brown, P.J, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SETDB1 in complex with a fragment candidate
To be published
|
|
5KCO
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy | | Descriptor: | DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5L7O
 
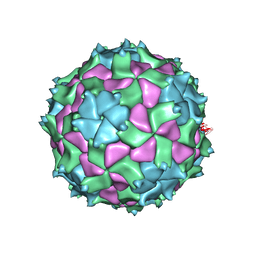 | | X-ray structure of Triatoma virus empty capsid | | Descriptor: | Capsid protein | | Authors: | Sanchez-Eugenia, R. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure of Triatoma virus empty capsid: insights into the mechanism of uncoating and RNA release in dicistroviruses.
J.Gen.Virol., 97, 2016
|
|
