4GPD
 
 | |
3E6D
 
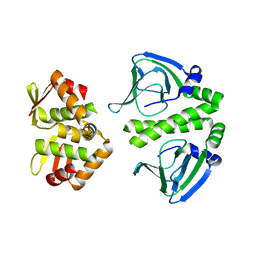 | | Crystal Structure of CprK C200S | | Descriptor: | Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3EGZ
 
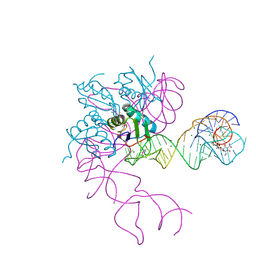 | | Crystal structure of an in vitro evolved tetracycline aptamer and artificial riboswitch | | Descriptor: | 7-CHLOROTETRACYCLINE, MAGNESIUM ION, Tetracycline aptamer and artificial riboswitch, ... | | Authors: | Xiao, H, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch
Chem.Biol., 15, 2008
|
|
3E5E
 
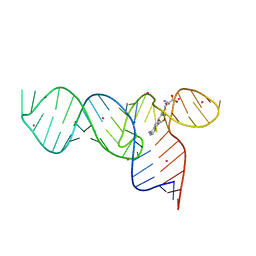 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2MHI
 
 | |
3EYA
 
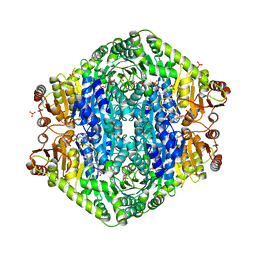 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EY9
 
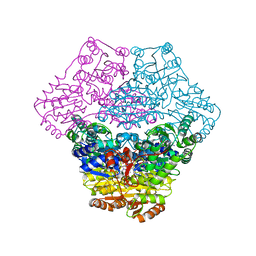 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate dehydrogenase [cytochrome], ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2N8T
 
 | |
3FSM
 
 | |
2ORH
 
 | |
2ORF
 
 | |
6VWB
 
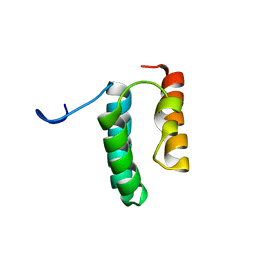 | | Solution structure of the N-terminal helix-hairpin-helix domain of human MUS81 | | Descriptor: | Crossover junction endonuclease MUS81 | | Authors: | Payliss, B, Houliston, S, Lemak, A, Arrowsmith, C.H, Wyatt, H.D.M. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease
Cell Rep, 41, 2022
|
|
3DH4
 
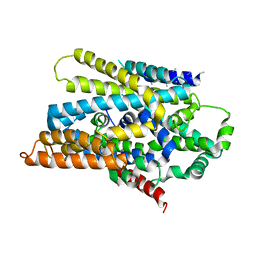 | | Crystal Structure of Sodium/Sugar symporter with bound Galactose from vibrio parahaemolyticus | | Descriptor: | ERBIUM (III) ION, SODIUM ION, Sodium/glucose cotransporter, ... | | Authors: | Abramson, J, Faham, S, Cascio, D. | | Deposit date: | 2008-06-16 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport.
Science, 321, 2008
|
|
3F5H
 
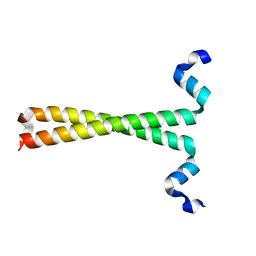 | | Crystal structure of fused docking domains from PikAIII and PikAIV of the pikromycin polyketide synthase | | Descriptor: | SODIUM ION, Type I polyketide synthase PikAIII, Type I polyketide synthase PikAIV fusion protein | | Authors: | Buchholz, T.J, Geders, T.W, Bartley, F.E, Reynolds, K.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains.
Acs Chem.Biol., 4, 2009
|
|
3FFO
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-2)man | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, Adhesin, ... | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
2ORG
 
 | |
4LYT
 
 | |
2HPR
 
 | |
2PAW
 
 | | THE CATALYTIC FRAGMENT OF POLY(ADP-RIBOSE) POLYMERASE | | Descriptor: | POLY(ADP-RIBOSE) POLYMERASE | | Authors: | Ruf, A, Schulz, G.E. | | Deposit date: | 1997-11-25 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling.
Biochemistry, 37, 1998
|
|
2PAX
 
 | |
4MDH
 
 | |
2KP7
 
 | | Solution NMR structure of the Mus81 N-terminal HhH. Northeast Structural Genomics Consortium target MmT1A | | Descriptor: | Crossover junction endonuclease MUS81 | | Authors: | Laister, R.C, Wu, B, Lemak, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-07 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and DNA binding mode of the Mus81 n-terminal helix-hairpin-helix domain
To be Published
|
|
2KXR
 
 | | ZO1 ZU5 domain MC/AA mutation | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
3Q8K
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with product 5'-flap DNA, SM3+, and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
