6TM3
 
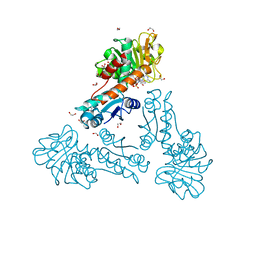 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in a close conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 1,2-ETHANEDIOL, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6BU3
 
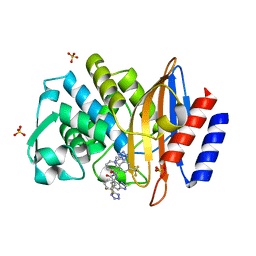 | |
3L1B
 
 | | Complex Structure of FXR Ligand-binding domain with a tetrahydroazepinoindole compound | | Descriptor: | 1-methylethyl 8-fluoro-1,1-dimethyl-3-{[4-(3-morpholin-4-ylpropoxy)phenyl]carbonyl}-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Farnesoid X receptor | | Authors: | Xu, W, Lundquist, J.T. | | Deposit date: | 2009-12-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Physiochemical Properties of the Tetrahydroazepinoindole Series of Farnesoid X Receptor (FXR) Agonists: Beneficial Modulation of Lipids in Primates.
J.Med.Chem., 53, 2010
|
|
6CW7
 
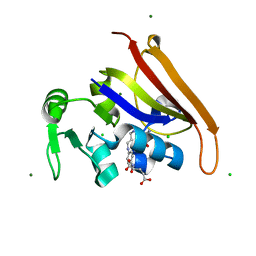 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
4A99
 
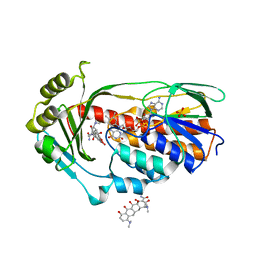 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH MINOCYCLINE | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2011-11-25 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4RPL
 
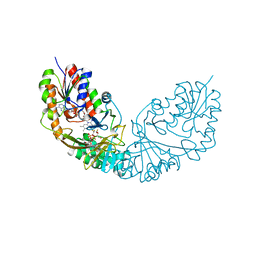 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4QPC
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ETS
 
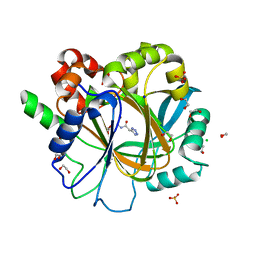 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
4LKV
 
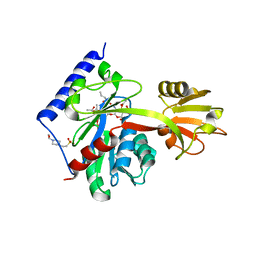 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5109 Å) | | Cite: | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
6ETT
 
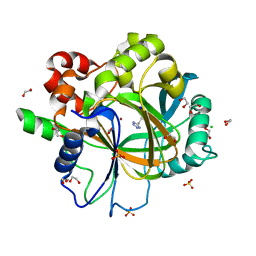 | | Crystal structure of KDM4D with tetrazole compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETV
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETU
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
5FHM
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-(aminomethyl)benzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at resolution 1.55 A resolution | | Descriptor: | (2~{S})-3-[5-[2-[[3-(aminomethyl)phenyl]methyl]-1,2,3,4-tetrazol-5-yl]-3-oxidanyl-1,2-oxazol-4-yl]-2-azanyl-propanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Al-musaed, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
6ETW
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 3 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
4PAA
 
 | | Crystal structure of the mature form of rat DMGDH complexed with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dimethylglycine dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Newcomer, M.E, Wagner, C. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Folate in demethylation: The crystal structure of the rat dimethylglycine dehydrogenase complexed with tetrahydrofolate.
Biochem.Biophys.Res.Commun., 449, 2014
|
|
6ETG
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
4R8V
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FHN
 
 | |
4PAB
 
 | | Crystal structure of the precursor form of rat DMGDH complexed with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dimethylglycine dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Newcomer, M.E, Wagner, C. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate in demethylation: The crystal structure of the rat dimethylglycine dehydrogenase complexed with tetrahydrofolate.
Biochem.Biophys.Res.Commun., 449, 2014
|
|
4RPK
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galf | | Descriptor: | (2R,5S)-5-[(1R)-1,2-dihydroxyethyl]-3,3,4,4-tetrafluorotetrahydrofuran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
5UOY
 
 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 16j (6-(4-Methoxybenzyl)-9-((tetrahydro-2H-pyran-4-yl)methyl)-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-methoxyphenyl)methyl]-9-[(oxan-4-yl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
6FFF
 
 | | Human BRD2 C-terminal bromodomain with (S)-5-(1-acetyl-2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-1,2,3,4-tetrahydroquinoxalin-6-yl)pyrimidine-2-carboxamide | | Descriptor: | (S)-5-(1-acetyl-2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-1,2,3,4-tetrahydroquinoxalin-6-yl)pyrimidine-2-carboxamide, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chung, C. | | Deposit date: | 2018-01-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Tetrahydroquinoxalines as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the Second Bromodomain.
J.Med.Chem., 61, 2018
|
|
6FFG
 
 | | Human BRD2 C-terminal bromodomain with (S)-1-(2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-6-(1,2,3,6-tetrahydropyridin-4-yl)-3,4-dihydroquinoxalin-1(2H)-yl)ethanone | | Descriptor: | (S)-1-(2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-6-(1,2,3,6-tetrahydropyridin-4-yl)-3,4-dihydroquinoxalin-1(2H)-yl)ethanone, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chung, C. | | Deposit date: | 2018-01-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Tetrahydroquinoxalines as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the Second Bromodomain.
J.Med.Chem., 61, 2018
|
|
4TT8
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TTS
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
