6M6A
 
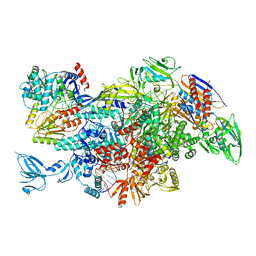 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6YXX
 
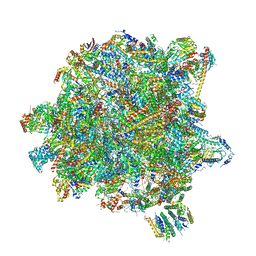 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6M6B
 
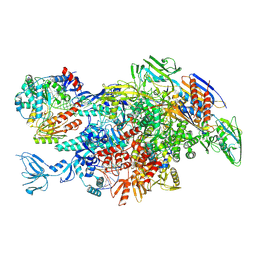 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6KYV
 
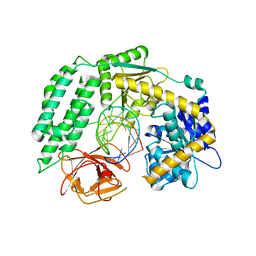 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
6ZQE
 
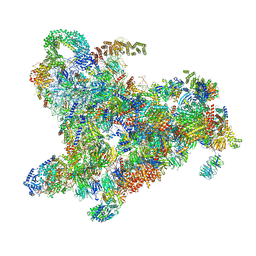 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQG
 
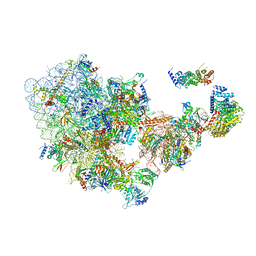 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6ZQD
 
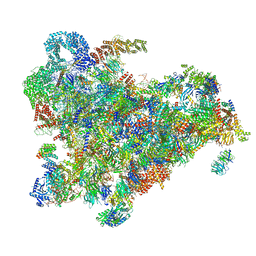 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQF
 
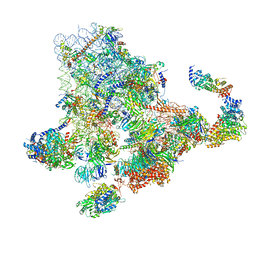 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6LQS
 
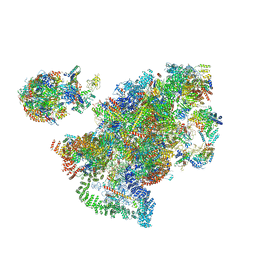 | |
6XEO
 
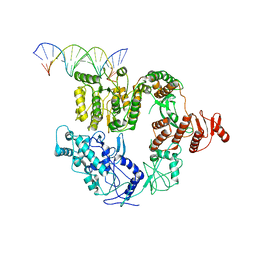 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
6Y50
 
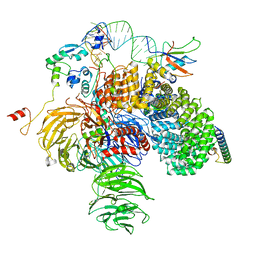 | | 5'domain of human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
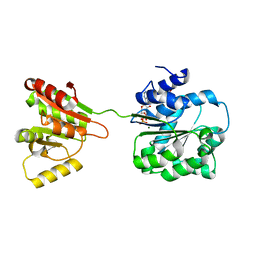 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6Y53
 
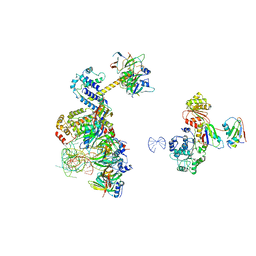 | | human 17S U2 snRNP low resolution part | | Descriptor: | HIV Tat-specific factor 1, Probable ATP-dependent RNA helicase DDX46, Small nuclear ribonucleoprotein E, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6Y5Q
 
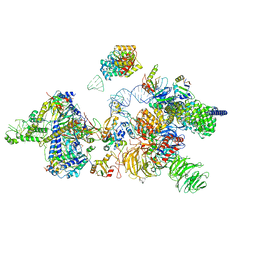 | | human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6YHR
 
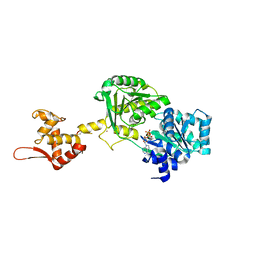 | | Crystal structure of Werner syndrome helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Werner syndrome ATP-dependent helicase, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the helicase core of Werner helicase, a key target in microsatellite instability cancers.
Life Sci Alliance, 4, 2021
|
|
6SH7
 
 | |
6SH6
 
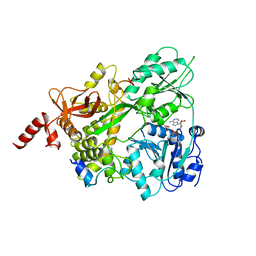 | | Crystal structure of the human DEAH-helicase DHX15 in complex with the NKRF G-patch bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jonas, S, Studer, M.K, Ivanovic, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for DEAH-helicase activation by G-patch proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UP4
 
 | |
6RM9
 
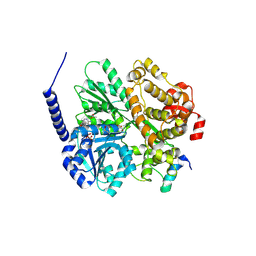 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMB
 
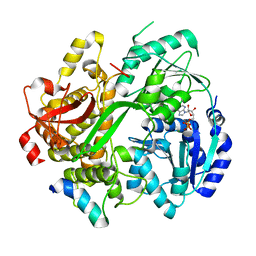 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
