3SNI
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3OKV
 
 | | Human Carbonic Anhydrase II A65S, N67Q (CA IX mimic) bound with 2-Ethylestrone 3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
1RRO
 
 | | REFINEMENT OF RECOMBINANT ONCOMODULIN AT 1.30 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, RAT ONCOMODULIN | | Authors: | Ahmed, F.R, Rose, D.R, Evans, S.V, Pippy, M.E, To, R. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refinement of recombinant oncomodulin at 1.30 A resolution.
J.Mol.Biol., 230, 1993
|
|
1OI9
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 6-CYCLOHEXYLMETHYLOXY-2-(4'-HYDROXYANILINO)PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
3SJT
 
 | | Crystal structure of human arginase I in complex with the inhibitor Me-ABH, Resolution 1.60 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxyhexyl](trihydroxy)borate | | Authors: | Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
2CJP
 
 | | Structure of potato (Solanum tuberosum) epoxide hydrolase I (StEH1) | | Descriptor: | 1,2-ETHANEDIOL, 2-PROPYLPENTANAMIDE, EPOXIDE HYDROLASE, ... | | Authors: | Mowbray, S.L, Elfstrom, L.T, Ahlgren, K.M, Andersson, C.E, Widersten, M. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Structure of Potato Epoxide Hydrolase Sheds Light on Substrate Specificity in Plant Enzymes.
Protein Sci., 15, 2006
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
2HU1
 
 | | Crystal structure Analysis of Hen Egg White Lyszoyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wine, Y, Cohen-Hadar, N, Freeman, A, Lagziel-Simis, S, Frolow, F. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis
Biotechnol.Bioeng., 98, 2007
|
|
1OIU
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
3DDG
 
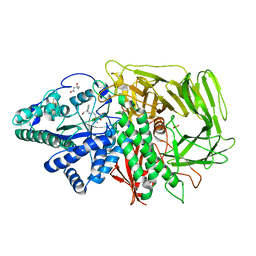 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) methylpyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)-1-methylpyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
1B9D
 
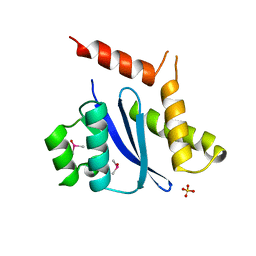 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
3SMR
 
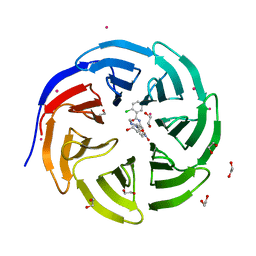 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3DI4
 
 | |
4CPO
 
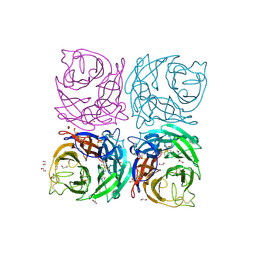 | | Structure of the Neuraminidase from the B/Lyon/CHU/15.216/2011 virus | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
1GR7
 
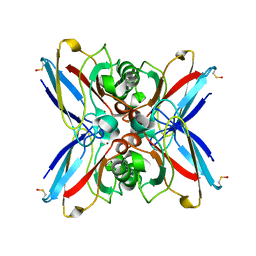 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
3G7B
 
 | |
1BHB
 
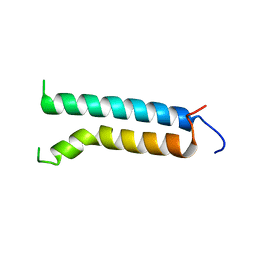 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
4HWV
 
 | |
2LS4
 
 | |
4HPJ
 
 | | Crystal structure of Tryptophan Synthase at 1.45 A resolution in complex with 2-aminophenol quinonoid in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
1BFD
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
1BHA
 
 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | Descriptor: | BACTERIORHODOPSIN | | Authors: | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | Deposit date: | 1993-10-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
2LS2
 
 | |
2HP2
 
 | | Inter-subunit signaling in GSAM | | Descriptor: | (4R)-5-AMINO-4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]PENTANOIC ACID, (4S)-4,5-DIAMINOPENTANOIC ACID, Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) hybrid-form, ... | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1S82
 
 | | PORCINE TRYPSIN COMPLEXED WITH BORATE AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, 1,3,2-DIOXABOROLAN-2-OL, CALCIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
