6JYL
 
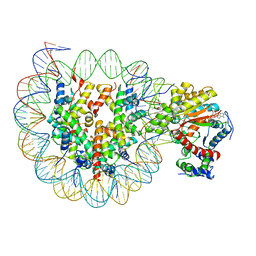 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3DX3
 
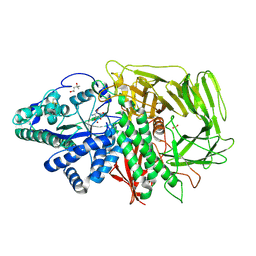 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1R,2R,3S,4R,5R)-5-aminocyclopentane-1,2,3,4-tetraol | | Descriptor: | (1R,2R,3S,4R,5R)-5-aminocyclopentane-1,2,3,4-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
6GNL
 
 | |
7XHL
 
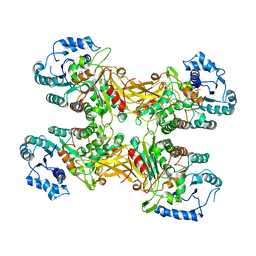 | | Complex structure of a Glucose 6-Phosphate Dehydrogenase from Zymomonas mobilis | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Glucose 6-Phosphate Dehydrogenase | | Authors: | Meng, D.D, Liu, M.X, Liu, W.D, You, C. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Coenzyme Engineering of Glucose-6-phosphate Dehydrogenase on a Nicotinamide-Based Biomimic and Its Application as a Glucose Biosensor
Acs Catalysis, 13, 2023
|
|
3DY5
 
 | | Allene oxide synthase 8R-lipoxygenase from Plexaura homomalla | | Descriptor: | Allene oxide synthase-lipoxygenase protein, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gilbert, N.C, Niebuhr, M, Tsuruta, H, Newcomer, M.E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A covalent linker allows for membrane targeting of an oxylipin biosynthetic complex.
Biochemistry, 47, 2008
|
|
6JYU
 
 | | Crystal structure of Human G6PD Canton | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Au, S.W.N. | | Deposit date: | 2019-04-28 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Human G6PD Canton
To Be Published
|
|
2GT3
 
 | |
6GOH
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
4QQS
 
 | | Crystal structure of a thermostable family-43 glycoside hydrolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoside hydrolase family 43, SODIUM ION | | Authors: | Hassan, N, Kori, L.D, Patel, B.K.C, Divne, C, Tan, T.C. | | Deposit date: | 2014-06-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution crystal structure of a polyextreme GH43 glycosidase from Halothermothrix orenii with alpha-L-arabinofuranosidase activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4R85
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine | | Descriptor: | 5-methylcytosine, Cytosine deaminase, FE (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine
To be Published
|
|
2HBT
 
 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | Authors: | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
4QSF
 
 | | CRYSTAL STRUCTURE of AMIDOHYDROLASE PMI1525 (TARGET EFI-500319) FROM PROTEUS MIRABILIS HI4320, A COMPLEX WITH BUTYRIC ACID AND MANGANESE | | Descriptor: | Amidohydrolase Pmi1525, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Xiang, D.F, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3E0L
 
 | | Computationally Designed Ammelide Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Murphy, P.M, Bolduc, J.M, Gallaher, J.L, Stoddard, B.L, Baker, D. | | Deposit date: | 2008-07-31 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Alteration of enzyme specificity by computational loop remodeling and design.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6K12
 
 | | Babesia microti lactate dehydrogenase apo form (BmLDH) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Long, Y. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6GFR
 
 | | cyanobacterial GAPDH with NAD | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4K57
 
 | |
6K0Z
 
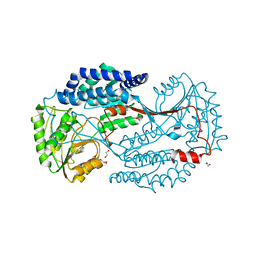 | | Substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GLYCEROL | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4967258 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
6GUO
 
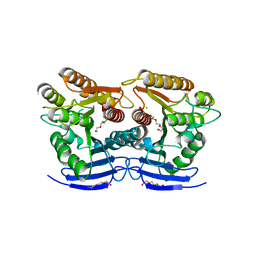 | | Siderophore hydrolase EstA from Aspergillus nidulans | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative siderophore-degrading esterase (Eurofung), ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4K22
 
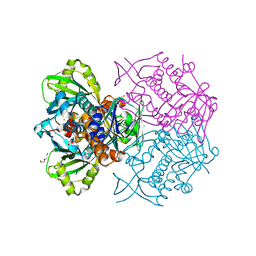 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
6GG7
 
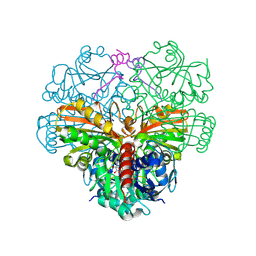 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3DX0
 
 | | Golgi alpha-Mannosidase II in complex with Mannostatin A at pH 5.75 | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
5XXQ
 
 | |
7XKA
 
 | | Structure of human beta2 adrenergic receptor bound to constrained epinephrine | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
5XYH
 
 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
6GI3
 
 | |
