2QXJ
 
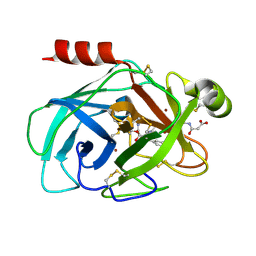 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone and Copper | | Descriptor: | COPPER (II) ION, Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1IYT
 
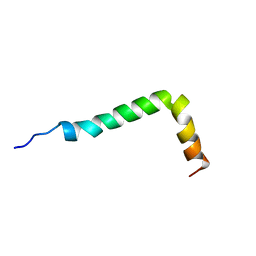 | | Solution structure of the Alzheimer's disease amyloid beta-peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Crescenzi, O, Tomaselli, S, Guerrini, R, Salvadori, S, D'Ursi, A.M, Temussi, P.A, Picone, D. | | Deposit date: | 2002-09-06 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain.
EUR.J.BIOCHEM., 269, 2002
|
|
5RFL
 
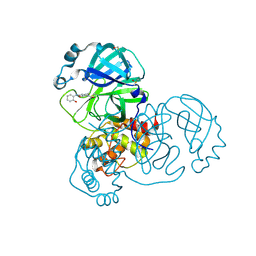 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102389 | | Descriptor: | 1-acetyl-N-(2-hydroxyphenyl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
3O9Z
 
 | | Crystal structure of the WlbA (WbpB) dehydrogenase from Thermus thermophilus in complex with NAD and alpha-ketoglutarate at 1.45 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Studies of WlbA: A Dehydrogenase Involved in the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid .
Biochemistry, 49, 2010
|
|
3G0Y
 
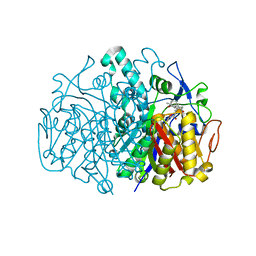 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | Descriptor: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3OCF
 
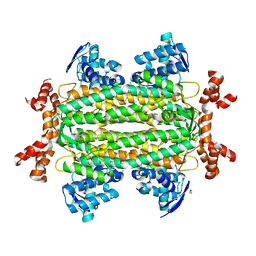 | |
1YXL
 
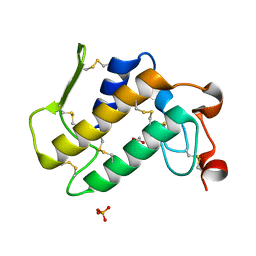 | | Crystal structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, R.K, Jabeen, T, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-02-22 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal Structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution
To be Published
|
|
1S6B
 
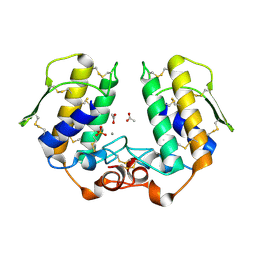 | | X-ray Crystal Structure of a Complex Formed Between Two Homologous Isoforms of Phospholipase A2 from Naja naja sagittifera: Principle of Molecular Association and Inactivation | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Jabeen, T, Sharma, S, Singh, R.K, Kaur, P, Singh, T.P. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a calcium-induced dimer of two isoforms of cobra phospholipase A2 at 1.6 A resolution.
Proteins, 59, 2005
|
|
1ZAA
 
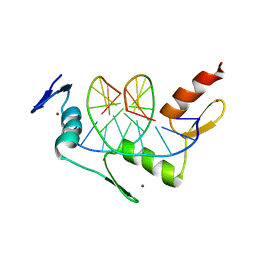 | |
2PCX
 
 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3SWR
 
 | |
1SDO
 
 | | Crystal Structure of Restriction Endonuclease BstYI | | Descriptor: | BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Vanamee, E.S, Edwards, T.A, Escalante, C.R, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2004-02-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of BstYI at 1.85 A Resolution: A Thermophilic Restriction Endonuclease with Overlapping Specificities to BamHI and BglII
J.Mol.Biol., 338, 2004
|
|
3OCI
 
 | | Crystal structure of TBP (TATA box binding protein) | | Descriptor: | 1,2-ETHANEDIOL, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Cui, S, Wollmann, P, Moldt, M, Hopfner, K.-P. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
2QXI
 
 | | High resolution structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QYV
 
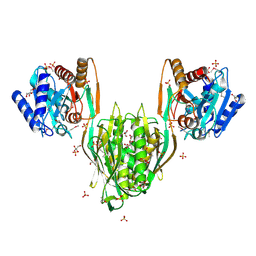 | |
2R1V
 
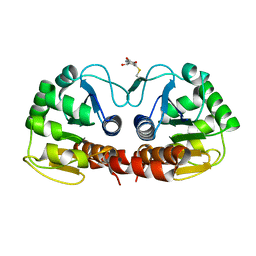 | |
2R1T
 
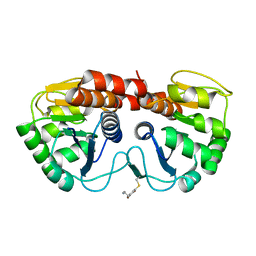 | |
2QAP
 
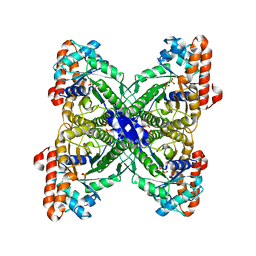 | |
2QKB
 
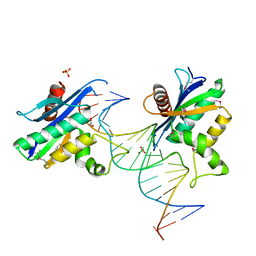 | | Human RNase H catalytic domain mutant D210N in complex with 20-mer RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DGP*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*DTP*DGP*DTP*DCP*DGP*DCP*DAP*DCP*DTP*DCP*DT)-3', 5'-R(*GP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2B55
 
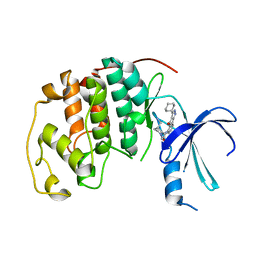 | | Human cyclin dependent kinase 2 (cdk2) complexed with indenopyraxole DIN-101312 | | Descriptor: | 2-(4-(AMINOMETHYL)PIPERIDIN-1-YL)-N-(3_CYCLOHEXYL-4-OXO-2,4-DIHYDROINDENO[1,2-C]PYRAZOL-5-YL)ACETAMIDE, Cell division protein kinase 2 | | Authors: | Muckelbauer, J. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and Evaluation of Indenopyrazoles as Cyclin-Dependent Kinase Inhibitors. 3. Structure Activity Relationships at C3
J.Med.Chem., 45, 2002
|
|
3OQ8
 
 | |
2QEY
 
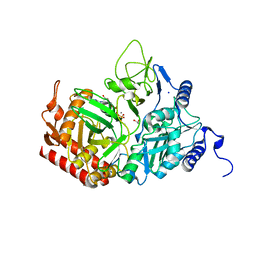 | | Rat cytosolic PEPCK in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
2QIS
 
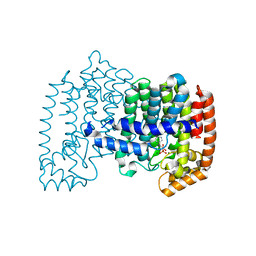 | | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Hozjan, V, Evdokimov, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate.
To be Published
|
|
2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
